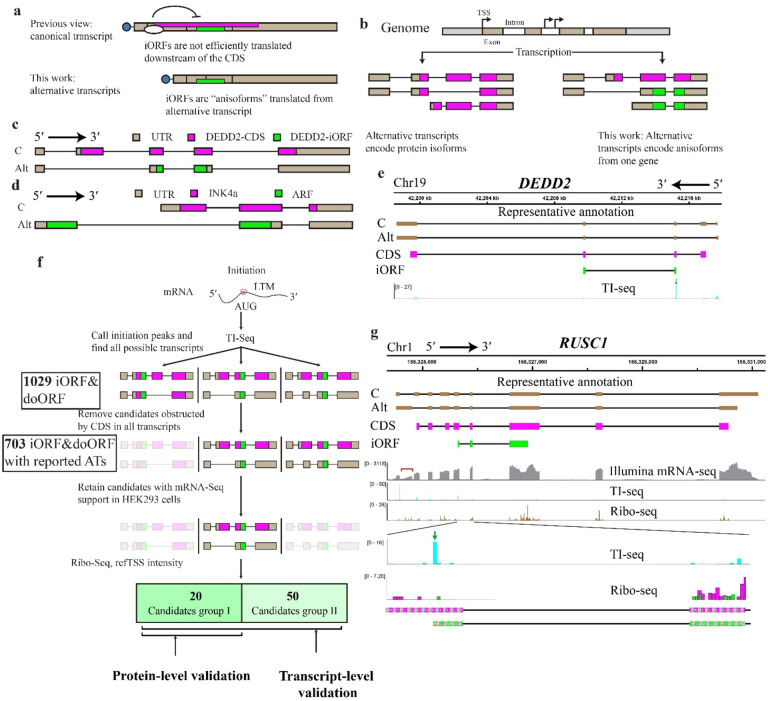
Fig. 1. Translation and global identification of anisoforms.
a, Frameshifted internal open reading frames (iORFs, green) have previously been mapped within the protein coding sequence (CDS, magenta) of annotated mRNAs (top, boxes represent exons), where their translation is likely inhibited by the upstream CDS start codon. We propose that iORFs are not translated from within annotated CDSs, but rather from alternative transcripts that lack the CDS start codon (bottom). If these alternative transcripts are currently known, they are likely annotated as non-coding variants based on the absence of the complete CDS. b, Pre-mRNA alternative splicing and alternative transcription sites (TSS) are known to generate alternative transcripts encoding protein isoforms with some constant, and some varied, domains. In contrast, alternative transcripts can also encode two entirely different proteins from the same gene (anisoforms) by including or skipping the translation initiation site of the CDS. c, Schematic of the previously reported DEDD2 gene, showing the canonical transcript and an alternatively spliced transcript. The alternative transcript lacks the second exon and the start codon for the canonical CDS, and only encodes the iORF. Transcript sequences were obtained from GENCODE. d, Schematic of the CDKN2A gene. The canonical transcript initiates from an upstream transcription start site (TSS) and encodes the INK4a protein. The alternative transcript skips the first exon, instead starting from an upstream TSS to produce the ARF protein in an alternative reading frame. e, Translation initiation (TI)-sequencing (seq) data (bottom track, blue peaks) supports expression of the previously reported DEDD2-SEP anisoform (iORF, green boxes, track 4), which overlaps the DEDD2 protein CDS (magenta boxes, track 3). A transcript variant specific for the DEDD2-SEP anisoform is currently annotated as non-coding (track 2), because it lacks exon 2 that contains the start codon of the DEDD2 CDS in the canonical transcript (track 1). f, Schematic of pipeline to identify candidate anisoforms from TI-seq data for experimental validation at the mRNA and/or protein level. The detailed workflow can be found in Method. ATs, alternative transcripts. g, Manual identification of an anisoform-encoding alternative transcript in the RUSC1 gene. Transcript 1 (C, track 1) represents the canonical transcript, while transcript 2 (Alt, track 2) represents an alternative transcript for the anisoform, both identified in GENCODE. The coding regions representing the RUSC1 CDS and the RUSC1 anisoform are schematized in track 3, magenta, and track 4, green, respectively. The mRNA-seq reads (track 5, gray) highlighted under the red bracket specifically support for the annotated alternative transcript 2. Previously published TI-seq signal (track 6, and zoom, track 8, cyan), provides evidence supporting the translation of anisoform. The green arrow indicates the peak for the start codon of anisoform in the TI-seq data. Reanalysis of previously reported Ribo-seq (tracks 7 and zoom, 9) supports low-resolution reads specific to the RUSC1 iORF reading frame (green) distinct from the CDS reading frame (magenta) within the overlapping region. Both TI-seq and Ribo-seq data were presented at the codon level.