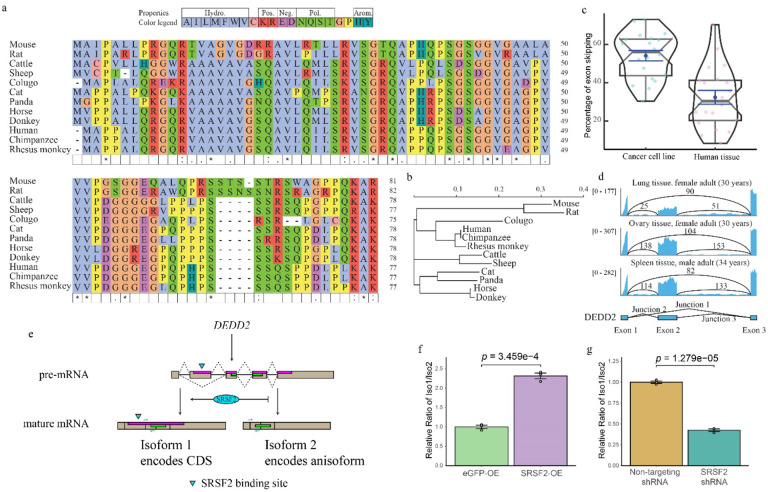
Fig. 6. Anisoform of DEDD2 is conserved and regulated by SRSF2.
a, Multiple sequence alignment of the DEDD2 anisoform sequence across twelve placental mammals, performed using seeded guide trees and HMM profile-profile techniques. Gaps in the alignment are marked with dashes. b, Phylogenetic tree constructed from the aligned DEDD2 anisoform sequences in a. Branch lengths indicate evolutionary distances (scale bar, arbitrary unit of evolutionary distance based on sequence divergence). c, Violin plots illustrating the percentage of DEDD2 exon 2 exclusion observed in RNA-seq data from ENCODE. The median values are indicated by horizontal blue lines. d, RNA-seq (ENCODE) read depth within exons 1 through 3 and counts of reads spanning splice junctions (exon 1–2, 2–3, and 1–3) in the DEDD2 gene, from three normal human tissues. e, Diagram illustrating requirement of SRSF2 binding for exon 2 inclusion in the DEDD2 mRNA. f and g. qRT-PCR measurement of the relative amounts of transcript variant 1 (exon 2 included) and 2 (exon 2 excluded) of the DEDD2 gene upon f, overexpression of SRSF 2 or the unrelated eGFP protein as a control, or g, shRNA-mediated silencing of SRSF2 vs a non-targeting shRNA control. p values, Welch two Sample t-test. Error bars represent the standard error of the mean (SEM) for n = 3.