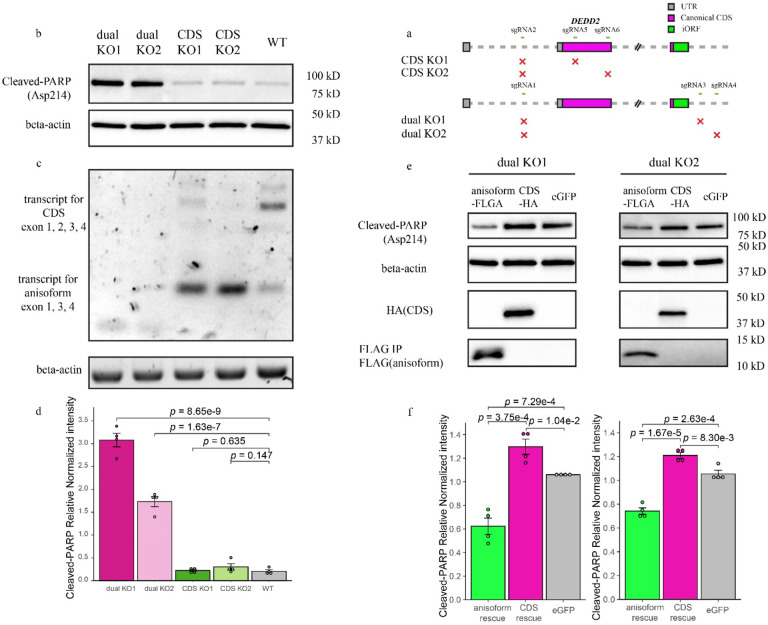
Fig. 7. DEDD2 anisoform is antiapoptotic.
a, Schematic representing the CRISPR/Cas9 editing strategy used to disrupt the DEDD2 gene to differentiate the expression and function of canonical CDS and anisoform. “CDS KO1” and “CDS KO2” used different sgRNA combinations to delete exon 2, which contains the start codon of the CDS and thus only deletes the DEDD2 CDS while leaving anisoform expression unchanged. “dual KO1” and “dual KO2” used different sgRNA combinations to delete exon 2, which contains the start codon of the CDS, and exon 3, which contains the start codon for the anisoform, thus abrogating expression of both DEDD2 and the anisoform. The gray squares represent UTRs, magenta bars denote the canonical CDS, and the green bar indicates the anisoform. KO validation can be found in Extended Data Fig. 9a and 9b. b, Western blot analysis of cleaved PARP (Asp214), an apoptosis marker, in wild-type, dual KO strains 1 and 2, and CDS-only KO 1 and 2. In all apoptosis assays, equal numbers of cells were seeded for each cell line, and apoptosis was induced with 200 ng/ml FasL for 4 hours followed by lysis and Western blotting. Beta-actin was a loading control. Results for additional independent clonal lines derived from each KO can be found in Extended Data Fig. 9c. c, RT-PCR and agarose gel analysis to detect altered transcript isoform production as a result of DEDD2 gene editing. RT-PCR of beta-actin was a loading control. d, Quantitation of normalized PARP cleavage with representative data shown in b. For all Western blot quantitation, cleaved PARP intensities were first normalized by the sum of replicates for each condition and then further normalized against actin levels to control for variations in protein loading and transfer across samples. Individual data points on the bars represent measurements from separate experimental replicates. All p values by two-sided, two-sample Student’s t-test. Error bars represent the standard error of the mean (SEM). n = 4. e, Western blot analysis of apoptosis marker cleaved PARP after rescue of dual knockout (KO) cell lines with either DEDD2 canonical protein or anisoform. Each clonal dual KO cell line was stably transfected via lentivirus with constructs encoding either the anisoform (FLAG-tagged), the canonical CDS (HA-tagged), or eGFP as a control. Beta-actin served as the loading control to ensure equal protein loading across samples. f, Quantitation of apoptosis marker in various cell lines (representative data in e) across four biological replicates as described in d.