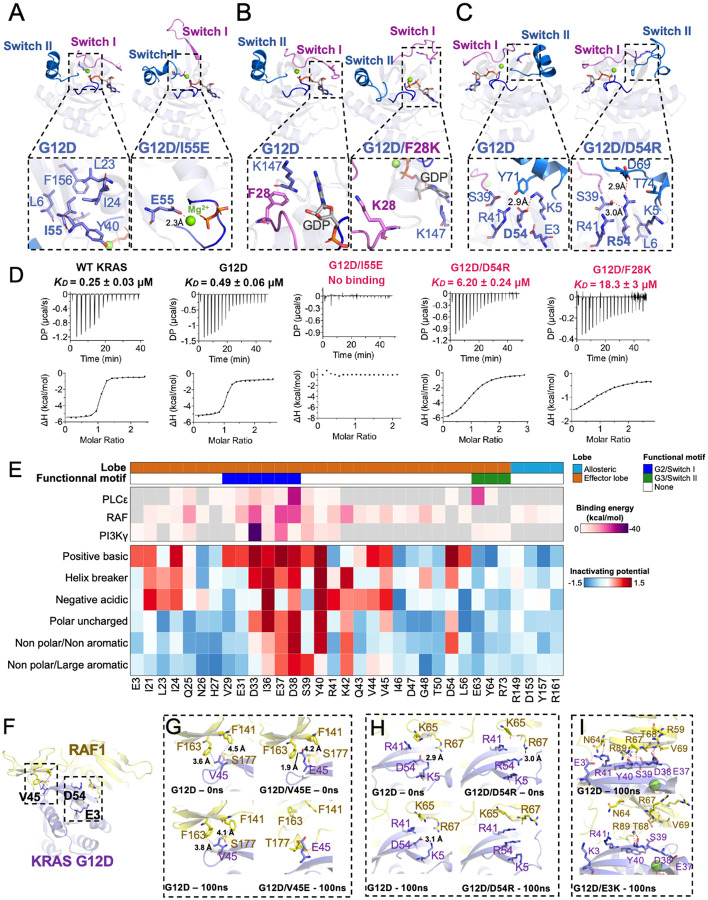
Figure 3. Structural insights and mutational tolerance profiles uncover KRASG12D inactivation mechanisms by allosteric and orthosteric impacts on switch-I and -II conformations.
Structural comparison of GDP-bound KRASG12D with (A) G12D/I55E, (B) G12D/F28K, and (C) G12D/D54R shows conformational changes in switch-I and -II caused by suppressor mutation. (D) Binding affinities (KD measured by isothermal titration calorimetry) for relevant KRASG12D inactivating mutants against effector RAF1-RBD, with inactivating mutants labeled in red. (E) Heatmap of KRAS effector binding residue interaction energy predicted by Amber10 force-field-based energy calculation (top) and average LFC of residues that have been grouped according to biophysical characteristics, including negative charge (D/E), positive charge (K/R), hydrophobic-aromatic (F/W/Y/H), hydrophobic-small (G/A/V/L/I/M), polar uncharged (S/T/C/Y/N/Q), and helix breaker (P/G). (F) Global structural view of KRAS and RAF1(RBD-CRD) with KRASG12D residues involved in direct RAF1 binding - V45, and proximal residues - E3 and D54 (stick representation) (PDB: 6XHB). Enlarged view of the KRAS-RAF1(RBD-CRD) interaction interface comparing KRASG12D against (G) V45E, (H) D54R, and (I) E3K.