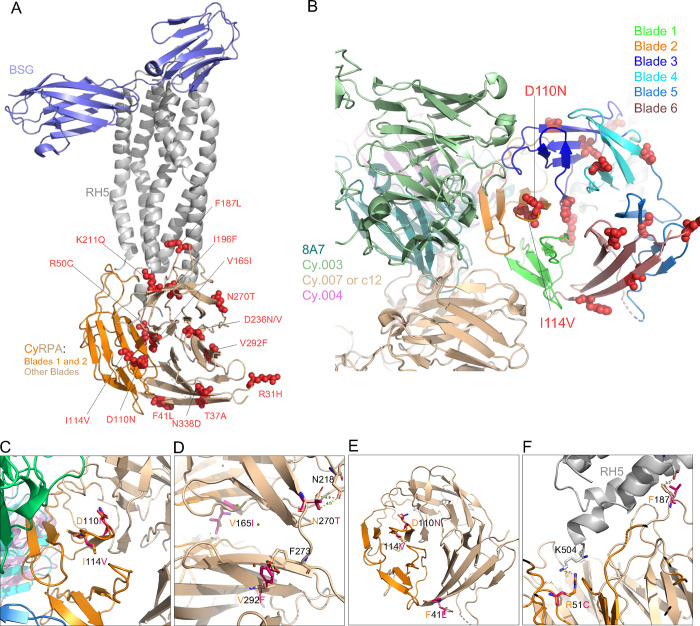
Figure 2: Structure-function predictions for the novel SNPs identified in CyRPA.
The complete structure was obtained by superimposing the structure of PfCyRPA in complex with PfRH5 (PDB id: 6MPV), PfRH5 bound to its ligand Basigin (PDB id: 4U0Q) and of known monoclonal antibodies Fab regions 8A7(PBD:5TIH), Cy.003 (PBD:7PI2), Cy.004 (PBD:7PHW) and Cy007/c12 (PBD:7PHV). Both CyRPA blades and individual antibodies are color coded. (A) The location of SNPs within the BSG–RH5–CyRPA complex. The complex construction was achieved by superimposing the RH5 of the RH5–BSG complex (PDB ID: 4U0Q) onto the RH5–CyRPA complex (PDB ID: 6MPV). BSG and RH5 are depicted in light blue and grey, respectively. CyRPA is represented in a wheat color, while blade 1 and blade 2 are indicated in dark orange. (B) shows the distribution of the SNPs across PfCyRPA blades and their position relative to the monoclonal antibody binding epitopes. BSG and PfRH5 ribbons are shown by light blue and grey, while the different blades of PfCyRPA are depicted in different colors. Ribbons of the 8A7, Cy.003, Cy.004 and Cy.007/c12 R5.011 are respectively shown dark green, light green, indigo and light marron. Pymol version 2.3.2 was used to predict the effect and to plot the structural location of each SNPs. (C) The positions of D110N and I114V relative to monoclonal antibodies (mAbs). Antibodies Cy.003, Cy.004, Cy.007, and 8A7 are represented in green, beige, magenta, and dark turquoise, respectively. (D) Structural modelling revealed that SNPs V165Y, N270T, and V292F influence the conformation of CyRPA. Small red plates signify the potential steric hindrances. (E) This panel highlights the SNPs that might confer resistance to antibodies, including F41L, D110N, and I114V. (F) SNPs R50C and F187L are shown to directly interact with PfRH5.