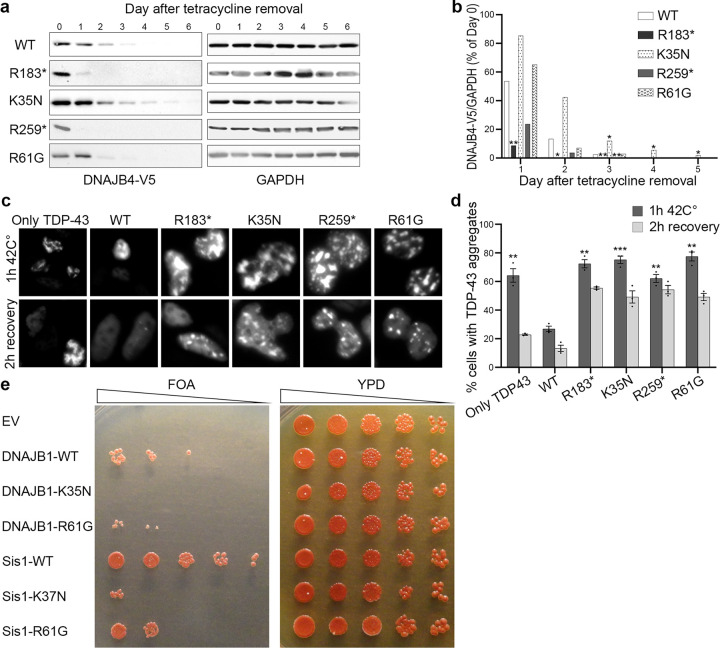
Figure 4.
DNAJB4 variants have a loss-of-function effect. (a) Tetracycline-regulated isogenic 293 cell lines were developed to express V5-tagged DNAJB4 (wild type, R183*, K35N, R259*, R61G). After induction with tetracycline for 24 hours, the cells were harvested at various times postinduction. Protein lysates were analyzed by immunoblotting for V5 and GAPDH, with GAPDH used as a loading control. (b) Densitometric analysis of V5-DNAJB4 normalized to GAPDH from three independent experiments, with the initial day’s value as a reference. Statistical significance was determined by comparing each variant to the WT on each day via a t test with Bonferroni correction for multiple comparisons (*P ≤ 0.0125, **P < 0.0025). (c) Representative fluorescence microscopy images displaying mCherry-tagged TDP-43 in HeLa cells post heat shock. (d) Quantification of cells displaying TDP-43 nuclear inclusions. n = 200–300cells analyzed per condition; the experiment was conducted 3 times. Statistical analysis was conducted by comparing the percentage of cells with TDP-43 aggregates after 1 hour at 42°C to that of WT cells via a t test with Bonferroni correction for multiple comparisons (**P < 0.002, ***P < 0.0002). (e) Yeast colonies lacking Sis1 were supplemented with either an empty vector (EV), wild-type DNAJB1 (DNAJB1-WT), DNAJB1-K35N, Sis1-WT, or the Sis1-K37N variant and then spotted on FOA media (left panel). The corresponding colonies on full media (YPD) are shown in the right panel. Data from five independent experiments were collected for each condition. EV, empty vector; WT, wild type.