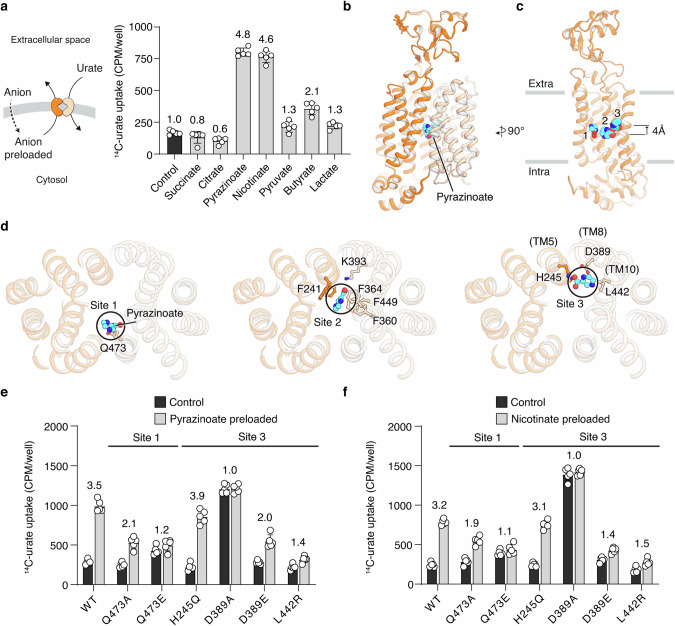
Fig. 4. Recognition of counter anion pyrazinoate by URAT1.
a Urate uptake of cells preloaded with various organic anions. The potentiation in urate uptake by anion preloading is presented as fold changes. Uptake buffer: 20 mM HEPES pH 7.4, 125 mM sodium chloride, and 5.6 mM glucose. Graph shows individual data points, mean ± SD; n = 5 biological replicates. b Pyrazinoate-bound URAT1 in an inward-facing conformation. c Three different pyrazinoate-binding sites in URAT1. For clarity, only the N-domain is shown. d Detailed interactions between pyrazinoate and URAT1, depicted in a top view looking down from the extracellular side. e Pyrazinoate potentiation of urate uptake. URAT1 variants with mutations in Sites 1 and 3 were examined. The potentiation in urate uptake by pyrazinoate preloading is presented as fold changes. Same buffer as in a. The graph shows individual data points, mean ± SD; n = 5 biological replicates. f Nicotinate potentiation on urate uptake. URAT1 variants with mutations in Sites 1 and 3 were examined. The potentiation in urate uptake by nicotinate preloading is presented as fold changes. The graph shows individual data points, mean ± SD; n = 5 biological replicates; same buffer as in a.