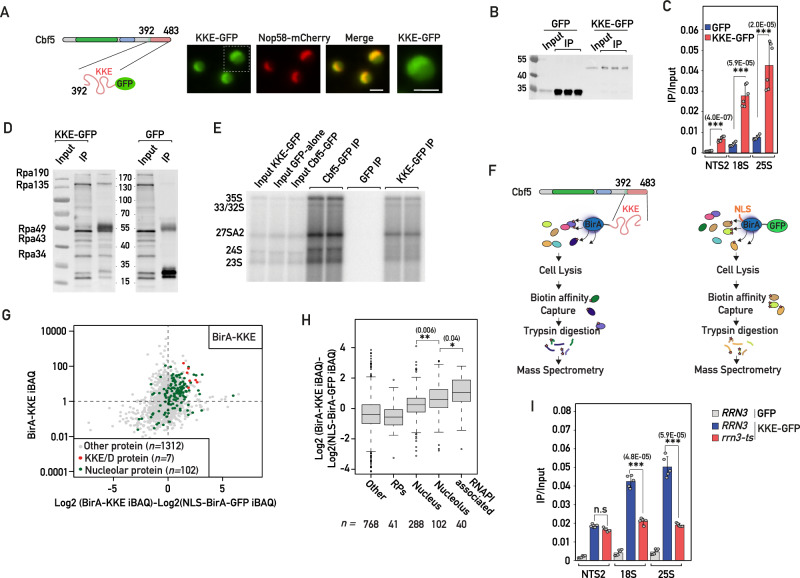
Fig. 4. The KKE/D domain is sufficient to promote efficient targeting close to transcribed rDNA genes.
A Strains expressing Nop58-mCherry and the KKE/D-GFP (KKE-GFP) construct were grown exponentially, and cells were analyzed by fluorescence microscopy. Zooms of the GFP signals are shown. Scale bars = 2 μm. B Western blot analysis using anti-GFP antibodies showing the IP efficiencies of GFP or KKE/D-GFP (KKE-GFP) in the ChIP-qPCR experiments shown in (C). C GFP and KKE/D-GFP (KKE-GFP) occupancy on rDNA genes at 18S, 25S or intergenic (NTS2) regions in wild-type cells evaluated by ChIP-qPCR. Unpaired two-tailed t-test analysis was used for statistics. Data are presented as mean values ± SD. Significant differences are indicated by stars and with the p value. n = 5 biologically independent experiment. D KKE/D-GFP (KKE-GFP) or GFP were immunoprecipitated using anti-GFP antibodies from formaldehyde-treated cells. Anti-RNAPI antibodies detecting all subunits were used to assess the co-immunoprecipitation of RNAPI. n = 2 biologically independent experiments. E KKE/D-GFP (KKE-GFP), Cbf5-GFP or GFP were immunoprecipitated using anti-GFP antibodies under native conditions. The co-immunoprecipitation of rRNA precursors was assessed by Northern blotting using radiolabeled probe 23S.1 (Supplementary Data 8). n = 2 biologically independent experiments. F Schematic representation of the TurboID-based proximity labeling experiments. G Scatter plot showing the normalized abundance value (iBAQ, intensity-based absolute quantification) of each protein detected in the purification of biotinylated proteins from cells expressing the BirA-KKE bait plotted against the relative abundance of these proteins (log2-transformed enrichment) compared to their normalized iBAQ value in the control purification from cells expressing the NLS-BirA-GFP bait. KKE/D domain-containing proteins and nucleolar proteins are labeled. n is indicated for each category. Source data are provided as a Source Data file. H The proteins detected in the TurboID-based proximity labeling assay were classified into non-overlapping subsets including RPs, proteins associated with RNAPI, proteins reported to localize in the nucleus, in the nucleolus or in other cellular areas. The log2-transformed enrichment is given for each category. p values were calculated with unpaired two-tailed Welch’s t test. Significant differences are indicated by stars and with the p value. n is indicated for each category. I KKE/D-GFP (KKE-GFP) or GFP occupancy on rDNA genes at 18S, 25S or intergenic (NTS2) regions in wild-type (RRN3) or rrn3-8 (rrn3-ts) mutant cells grown at 37 °C. p values were calculated and indicated as in (C). n = 5 biologically independent experiments. Source data are provided as a Source data file.