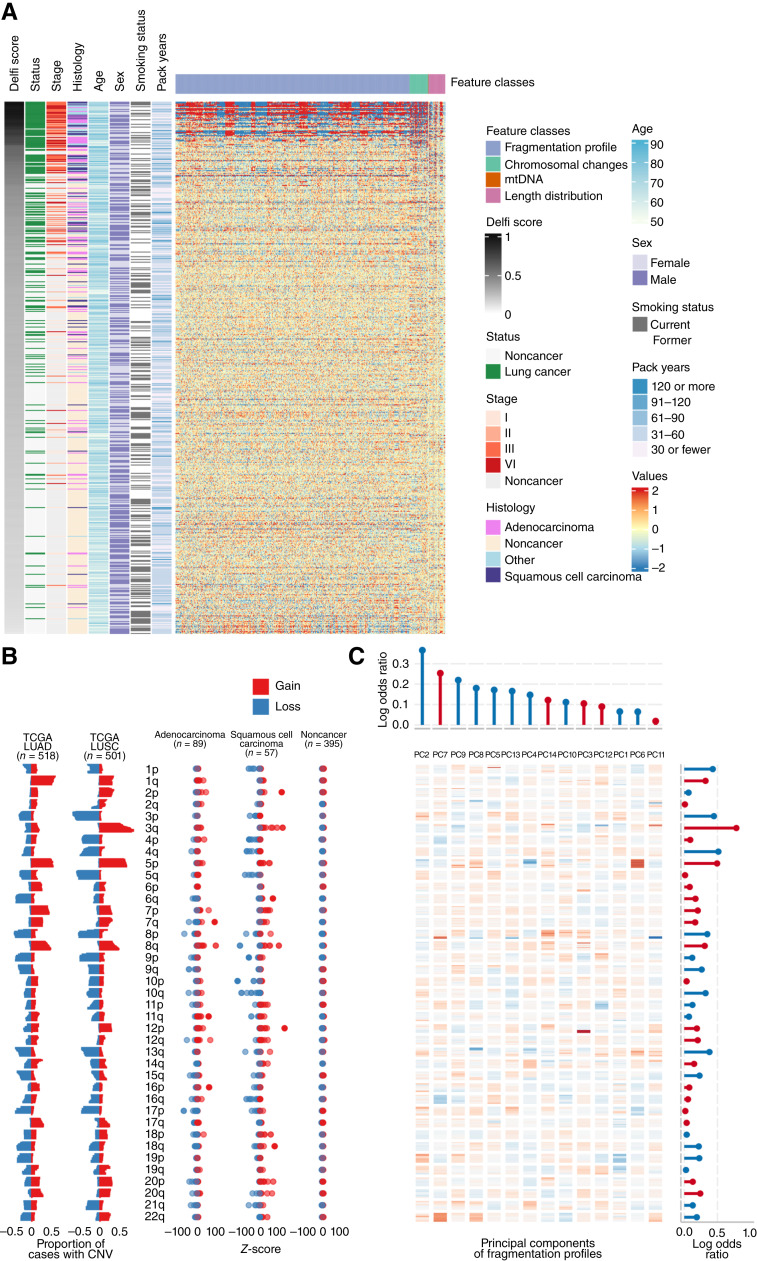
Figure 3.
High-dimensional fragmentation features reflect lung cancer biology and are incorporated in the machine learning classifier. A, Heatmap representation of the deviation of cfDNA fragmentation features across the genome for the classifier training set with lung cancer or noncancer individuals compared with the mean of classifier training noncancer individuals. Each row represents a sample, whereas columns show individual genomic features. The cross-validated DELFI score and clinical characteristics are indicated to the left of the fragmentation deviation heatmap. B, Left, TCGA-derived observations of chromosomal arm gains (red) and losses (blue) in lung adenocarcinoma (LUAD; n = 518) and squamous cell cancer tissues (LUSC; n = 501). Right, the observed chromosome arm gains (red) and losses (blue) in the classifier training individuals separated by histology. C, A heatmap representation of the principal component eigenvectors of the fragmentation profile features. Regression coefficients from the final classifier indicating how the principal components of the fragmentation profiles and z-scores of the chromosomal arms were combined are provided in the top and right margins of the heatmap, respectively. Positive values for the coefficients are represented in red, whereas negative values are represented in blue. Agreement across copy number chromosomal gains and losses in TCGA lung cancers, observed z-scores in the cfDNA of patients with lung cancer, and chromosome arm model coefficients reflect biologic consistency between chromosomal changes in lung cancer, cfDNA fragmentation profiles, and classifier features.