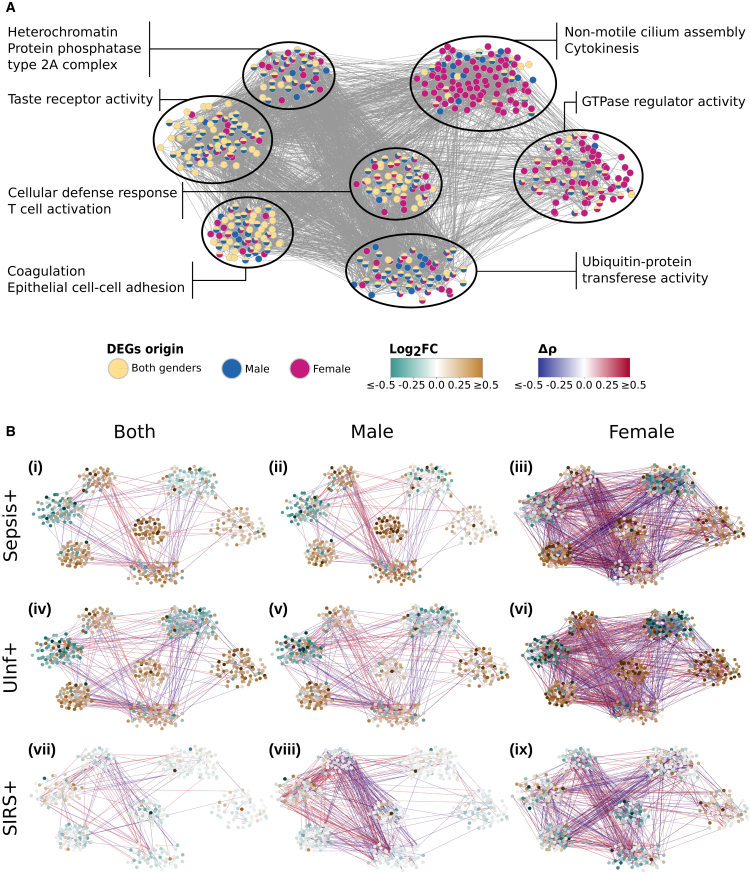
Figure 2.
Gene co-expression networks
(A) Visualization of merged co-expression networks based on annotated differentially expressed genes (DEGs) using preoperative samples with infection or non-infection postoperative outcome. Edges refer to Spearman correlations of ≥0.7 between genes in at least one sample set (all samples, only male, or only female samples). Node color indicates for which preoperative sample set the respective gene was significantly differentially regulated. Enriched Gene Ontology (GO) terms for identified modules of the co-expression network are indicated.
(B) Visualization of expression and correlation changes given preoperative samples with sepsis, UInf+, or SIRS+ postoperative outcome compared against SIRS− postoperative outcome using all, male-, or female-associated samples. Node colors indicate the log2 fold change of mean normalized gene expression between each sub-group (sepsis, UInf+, SIRS+) against SIRS− (control). Edge colors indicate the difference in gene-gene Spearman correlations between each sub-group against SIRS−. Spearman correlation differences ≤0.3 were omitted for clarity.