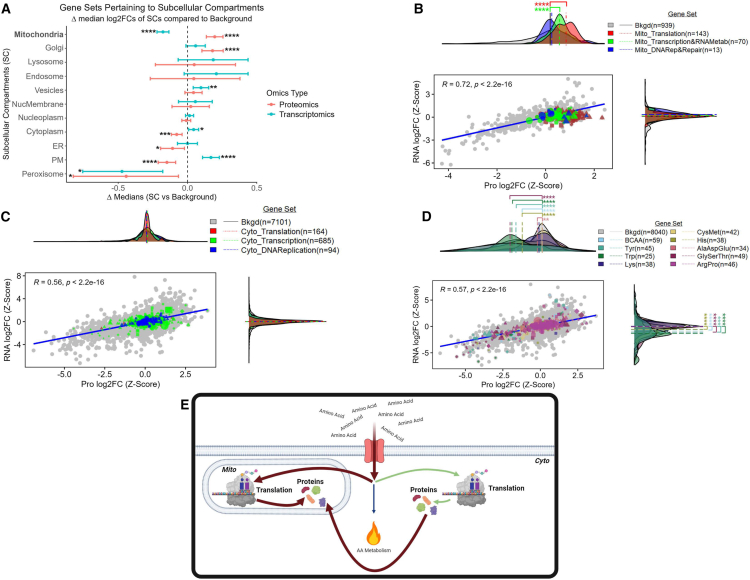
Figure 2.
Enhanced transcription and translation of mitochondrial genes
(A) Dot plot where each dot represents the difference (x axis) between the median log2FC (FLC vs. NML) of the background set and a gene set that represents a subcellular compartment (y axis). Error bars represent 95% confidence intervals.
(B–D) Comparison of log2FCs (FLC vs. NML; z-scaled) at the protein (x axis) and RNA (y axis) level. Density plots represent the distribution of the log2FCs associated with proteins (x axis) and transcripts (y axis). (E) Descriptive model of amino acid utilization in FLC. Gene sets were identified as members of (A) subcellular compartments, (B) the mitochondrial central dogma, (C) the cytoplasmic central dogma, and (D) amino acid metabolism. Background sets (bkgd) are (A and D) all genes identified in both the transcriptomic and proteomic datasets, (B) all genes related to the mitochondrion, and (C) all genes not related to the mitochondrion. (B–D) Lines associated with density plots represent the median. (A–C) MitoCarta3.0 and (A, C, and D) Protein Atlas were used to identify gene sets. (A–D) Gene sets (transcriptomic—n = 27 samples, proteomics—n = 23 samples) were compared to bkgd using Mann-Whitney (∗FDR-adj p < 0.05; ∗∗FDR-adj p < 0.01; ∗∗∗FDR-adj p < 0.001; ∗∗∗∗FDR-adj p < 0.0001). (B–D) For gene sets (excluding bkgd)—circles, genes with no significant log2FCs at either RNA or protein level; diamonds, genes with significant log2FCs at both the RNA and protein level; triangles, genes with significant log2FCs at the protein level only; squares, genes with significant log2FCs at the RNA level only; shape size, average expression values (from transcriptomic data). (A–D) Cyto, cytosol; Mito, mitochondria; DNARep&Repair, DNA replication and repair; BCAA, branched-chain amino acids; ER, endoplasmic reticulum; Lyso, lysosome; NucMem, nuclear membrane; Nucplasm, nucleoplasm; PM, plasma membrane; Perox, peroxisome.