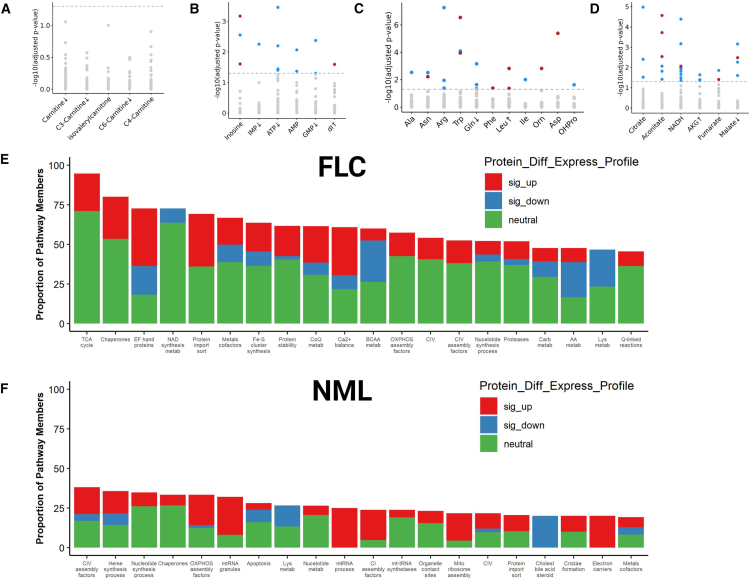
Figure 3.
Proteo-transcriptomic signature of mitochondrial respiration suggests an enhanced TCA cycle
(A–F) Concordance analysis in FLC patient tissue (n = 17 omics-matched sample pairs, 13 of which are FLC) between protein expression of (A–D) mitochondrial small-molecule transporters (data points) and metabolites (x axis labels) composed of (A) FA/carnitine conjugates, (B) nucleotides, (C) amino acids, and (D) TCA cycle intermediates. (A–D) Highlighted data points denote protein-metabolite pairs that are significantly (−log10(FDR-adj p) > 1.3) concordant (red) or discordant (blue) in FLC. Arrows adjacent to labels denote direction of significant alterations in the abundance of metabolites (x axis labels; FDR-adj p < 0.1) in FLC compared to NML. Gray data points indicate protein-metabolite pairs that were not statistically significant in concordance or discordance. Level of agreement between mitochondrial content (measured by Mitochondria Deep Tracker Red Fluorescent Intensity—MTDR F.I.) and protein abundance of all genes related to the mitochondria in (E) FLC (nmatchedpairs = 3; nrandompairs = 3) and (F) NML (nmatchedpairs = 4; nrandompairs = 2). Bars represent the top twenty mitochondrial pathways (x axis) with the highest percentage (y axis) of protein members whose abundance (proteomics) was significantly associated with mitochondrial content in (E) FLC and (F) NML. (E and F) Colors correspond to percentage of protein members that are significantly upregulated (sig_up, red), significantly downregulated (sig_down, blue), or unchanged (neutral, green) in FLC compared to NML. Protein_Diff_Express_Profile, protein differential expression profile; sig_up, significantly up; sig_down, significantly down; Carb, carbohydrate; BCAA, branched-chain amino acids; AA, amino acids; metab, metabolism; Cholest, cholesterol. (E and F) MitoCarta3.0 was used to identify gene sets.