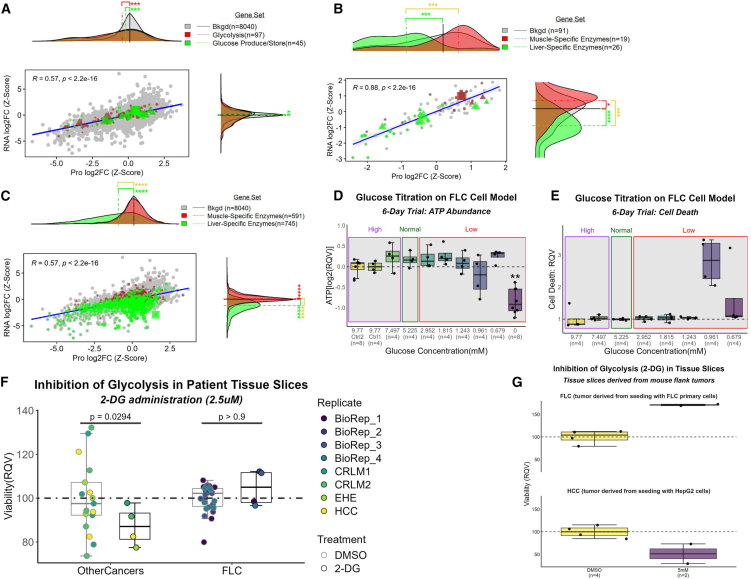
Figure 4.
Suggestive decoupling of glycolysis from pyruvate production
(A–C) Comparison of log2FCs (FLC vs. NML; z-scaled) at the protein (n = 23 samples) and RNA (n = 27 samples) level. Gene sets were identified that (a) participate in glycolysis as well as glucose production/storage and have (B and C) tissue specificity to liver or muscle. Background set (bkgd) is (B) all genes that participate in glycolysis and (A and C) all genes identified in both omics’ datasets. (A–C) Protein Atlas database was used to identify gene sets. Six-day glucose titration assay on FLC cell line measuring (D) ATP abundance (n = 8 samples for Ctrl 2 and zero concentration; n = 4 samples each for remaining concentrations) and (E) cell death (n = 4 samples per concentration).
(D–G) High, Normal, and Low indicate glucose concentrations above, within, and below normal physiological range, respectively. Inhibition of glycolysis via 2-deoxyglucose (2-DG) on fresh tissue slices from (F) patients afflicted with FLC (n = 4 biological replicates) or other cancers and (G) NOD scid gamma (NSG) mouse flank tumors seeded by either HepG2 or FLC primary cells. (F) BioRep, biological replicate; DMSO, control condition; CRLM, colorectal liver metastases; EHE, epithelioid hemangioendothelioma; HCC, hepatocellular carcinoma. (D–F) RQV, relative quantitative value. Mann-Whitney used for statistical assessment of (A–C) (∗FDR-adj p < 0.05; ∗∗FDR-adj p < 0.01; ∗∗∗FDR-adj p < 0.001; ∗∗∗∗FDR-adj p < 0.0001). (A–C) For gene sets (excluding bkgd), circles, genes with no significant log2FCs at either RNA or protein level; diamonds, genes with significant log2FCs at both the RNA and protein level; triangles, genes with significant log2FCs at the protein level only; squares, genes with significant log2FCs at the RNA level only; shape size, average expression values (from transcriptomic data).