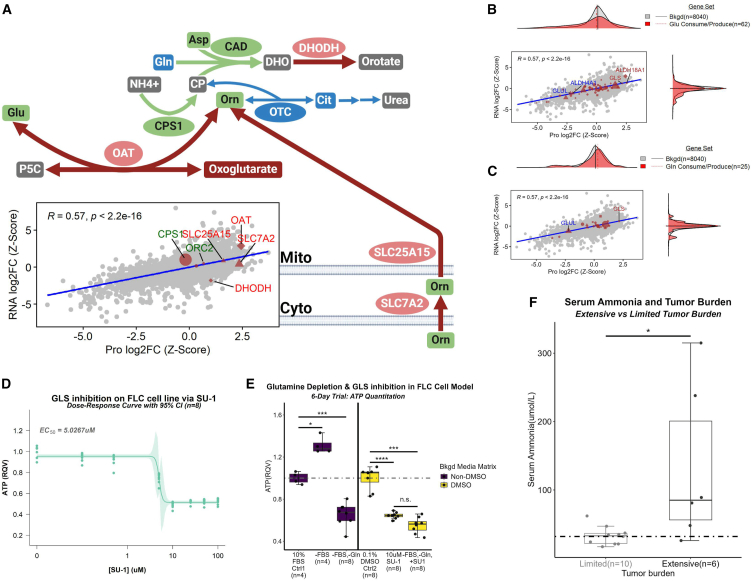
Figure 7.
Glutamine likely serves as a critical substrate for proline production in FLC
(A–C) (A) Schematic (top) and correlation plot (bottom) depicting OAT nexus in FLC. Coloration of labels in correlation plot and nodes in schematic correspond to proteins and metabolites with a significant increase (red) or decrease (blue) in log2FC in FLC (proteomics—n = 16 samples; metabolomics—n = 20 samples) compared to NML (proteomics—n = 7 samples; metabolomics—n = 6 samples). Green coloration indicates no significant alteration. Comparison of log2FCs (FLC vs. NML; z-scaled) at protein (n = 23 samples) and RNA (n = 27 samples) level for genes that participate in the consumption or production of (B) glutamate (Glu) or (C) glutamine (Gln).
(D) Dose-response curve of FLC cell viability with varying concentrations (n = 8 replicates per concentration) of SU-1, a pan-glutaminase inhibitor. EC50 = 5.0267 μM. Light-shaded green area around curve represents 95% confidence interval.
(E) FLC cell viability under glutamine depletion and/or GLS inhibition. Mann-Whitney was used for statistical analysis. ∗ FDR-adj p < 0.05; ∗∗∗ FDR-adj p < 0.001; ∗∗∗∗ FDR-adj p < 0.0001; n.s., not significant.
(F) Serum ammonia levels in patients afflicted with FLC comparing limited (n = 10) and extensive (n = 6) tumor burden using Mann-Whitney test (∗FDR-adj p < 0.05). JHU, John Hopkins University. Dotted line indicates the upper limit for serum ammonia. (A) For schematic, arrow size is directly associated with arrow color. Node size is arbitrary. Red, increase; green, neutral; blue, decrease; gray, no data; elliptical node, protein; square node, metabolite; arrow, reaction; broken arrows, series of reactions. (B and C) Human Metabolome Database was used to identify gene sets. (B and C) Gene sets were compared to bkgd using Mann-Whitney test (∗FDR-adjusted p < 0.05; ∗∗FDRFDR-adjusted p < 0.01; ∗∗∗adjusted p < 0.001; ∗∗∗∗adjusted p < 0.0001). (A–C) Background set (bkgd) is all genes identified in both omics’ datasets. For the gene sets (excluding bkgd), circles, genes with no significant log2FCs at either RNA or protein level; diamonds, genes with significant log2FCs at both the RNA and protein level; triangles, genes with significant log2FCs at protein level; squares, genes with significant log2FCs at RNA level; shape size, average gene expression values (from transcriptomic data). (E) Bkgd, Background; RQV, relative quantitative value; Ctrl, control; FBS, fetal bovine serum; Gln, glutamine.