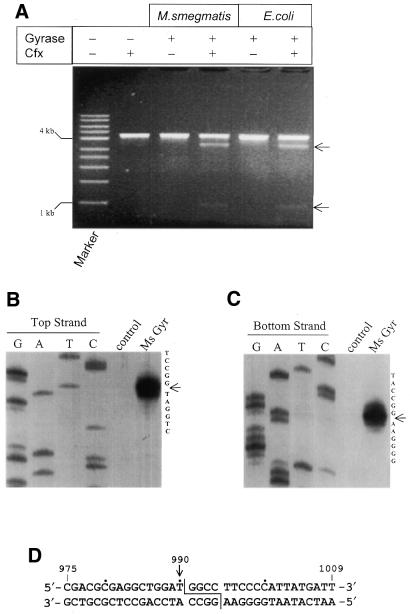
Figure 5.
DNA cleavage site mapping of M.smegmatis DNA gyrase. (A) A comparison of ciprofloxacin-induced DNA cleavage pattern of linearised pBR322 between E.coli and M.smegmatis DNA gyrases. Marker, a 1 kb DNA ladder (Sigma). Arrows indicate the cleaved products (0.9 and 3.4 kb). (B and C) Precise mapping of cleavage position. Cleavage site in the top (B) and bottom (C) strands of the 240 bp DNA fragment of pBR322 encompassing the SGS. Cleavage reactions were performed with labelled DNA fragments in the presence of ciprofloxacin (30 µg ml–1). G, A, T and C are sequencing lanes; lanes 5 and 6 are cleavage reactions in the absence and presence of M.smegmatis DNA gyrase, respectively. (D) Mycobacterial DNA gyrase cleavage positions in pBR322. Nucleotides are numbered showing the cleavage site position at 990.