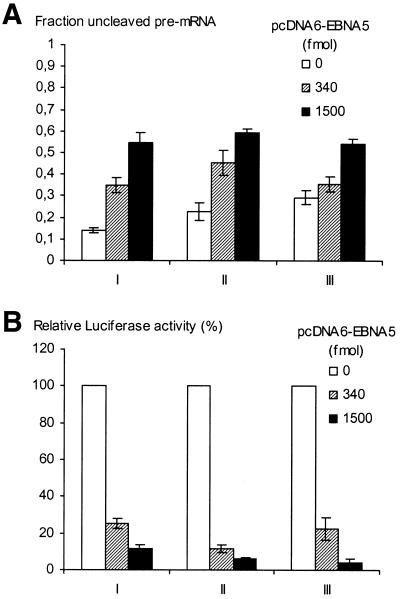
Figure 6.
EBNA5-induced inhibition of pre-mRNA processing depends on sequences in the reporter gene transcript. The reporter gene region of pCMVLuc encompassing the promoter (CMV), the luciferase-encoding sequence and the 3′-UTR [the SV40 late poly(A) signal] was amplified by nested PCR. The primers in the first round were 5′-AGTGCAAGTGCAGGTGCCAG-3′ (sense) and 5′-TGACTGGGTTGAAGGCTCTC-3′ (antisense) and in the second round 5′-GAGCTCTTACGCGTGCTAGC-3′ (sense) and 5′-TCTCAAGGGCATCGGTCGAC-3′ (antisense). The primer sequences were derived from pGL3-Basic vector (Promega) regions surrounding the reporter gene. Aliquots of 5 × 106 DG75 cells were co-transfected with circular pCMVLuc DNA (I), pCMVLuc DNA cleaved with BamHI and NheI to release the luciferase gene region (II) and the PCR-amplified DNA fragment of the luciferase gene region (III), respectively, and increasing amounts (0, 340 and 1500 fmol) of pcDNA6-EBNA5 DNA. (A) Total RNA was prepared and analysed for the presence of cleaved and uncleaved luciferase pre-mRNA using a RNase protection assay with a riboprobe comprising the SV40 3′-UTR cleavage signal. The relative levels of cleaved (C) and uncleaved (U) probe fragments were quantified by phosphorimaging. The results of five independent experiments are presented as the means ± SEM of the ratio between the uncleaved and the sum of cleaved and uncleaved fragments. (B) Analysis of the pCMVLuc reporter activity in the transfected cell extracts. The measured values for each set of experiments (I–III) are expressed as a percentage of the value obtained in the absence of EBNA5. The means ± SEM of five independent experiments are presented.