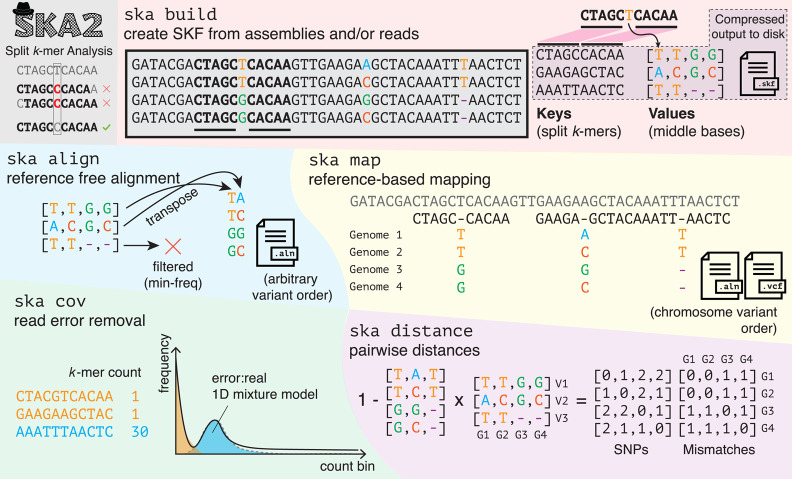
Figure 1.
Overview of functions and methods in SKA2. Split k-mers allow matching variant positions, whereas contiguous k-mers mismatch any variation. ska build creates split k-mer dictionaries from input sequence data. The example shows four sequences that are aligned and on the same strand for clarity, but in real input data, neither is necessary. Split k-mers are used as keys, and their middle bases are stored in lists. This dictionary is compressed using snappy to make split k-mer files (SKFs). ska align makes reference-free alignments with no coordinate system by writing out the middle bases, applying filters on the frequency of missing data, constant sites, and ambiguous sites. ska map makes reference-based mappings as ALN or VCF, with the same coordinate system as the reference. In both modes, the conserved sites are also written out but are not shown for clearer visualization. ska cov counts k-mers and fits a mixture model to find a threshold for count when using reads as input to ska build. ska distance calculates SNP distances and mismatches between samples by multiplying the middle base matrix by its transpose. The cluster_dists.py script can be run on this distance matrix to make phylogeny, single-linkage clusters with a provided threshold, and a Microreact visualization. Operations to merge, delete samples and split k-mers, and write out the contents of SKFs are also implemented but are not shown.