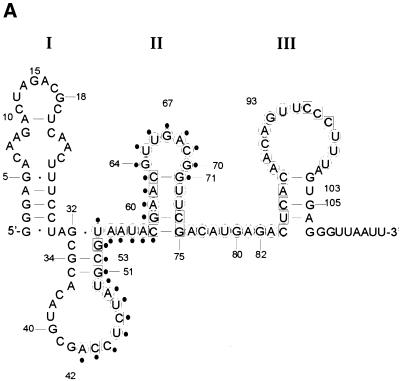
Figure 5.
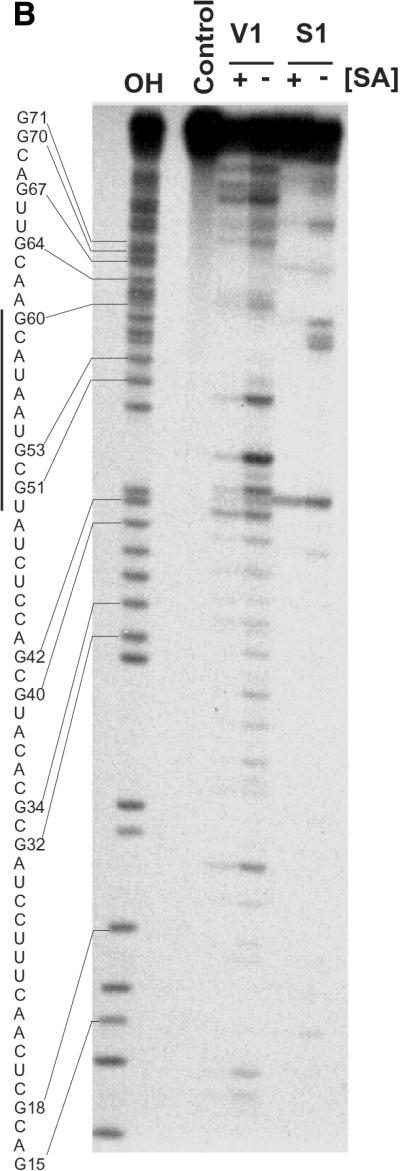
Solution structure of SA19 aptamer and SA footprinting. (A) Proposed secondary structure of SA19 aptamer as deduced from solution probing data and predictions with STAR software package. Reactive nucleotides towards DMS (C at N3), CMCT (U at N3, G at N1) and kethoxal (G at N1, N2) are encircled, and non-reactive nucleotides are boxed. No symbol is for not determined. The protected area is delimitated by black dots. (B) Autoradiogram of a 18% polyacrylamide/8 M urea gel, showing digestion products of 5′-end-labeled SA19 with RNase V1 and nuclease S1 in the presence (+) or absence (–) of SA; the major protected area is shown by a vertical line. Lane OH is the ladder from partially alkaline hydrolyzed SA19. The control lane corresponds to the 5′-end-labeled SA19 incubated in the presence of SA but in the absence of any nucleases. The gaps in the OH ladder are indicative of 2′-F-pyrimidines.