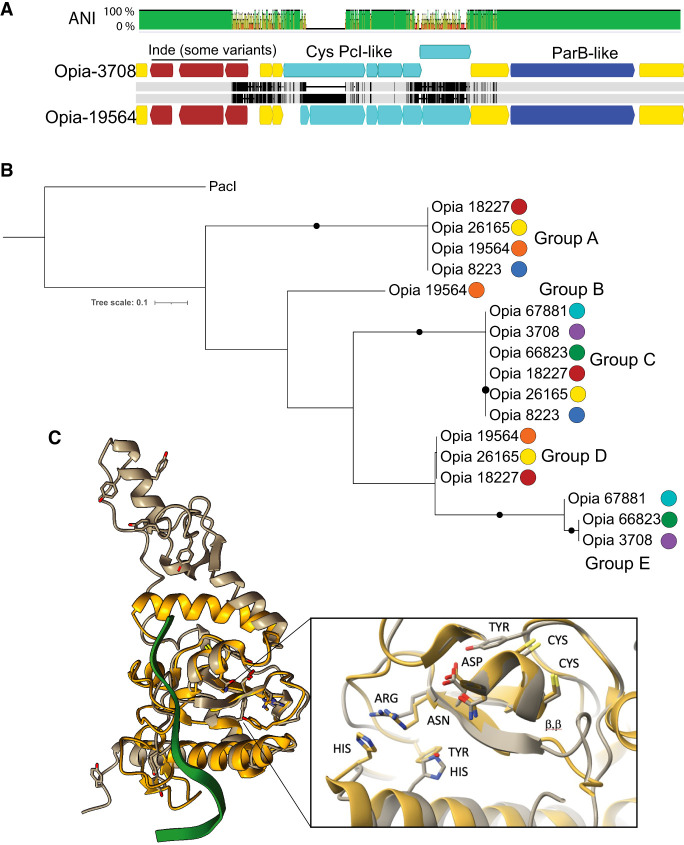
Figure 6.
Sequence variation in the cysteine-rich proteins of Opia viruses and comparison to Pacl, a rare-cutting HNH restriction endonuclease. (A) The aligned ∼5.6 kbp variable region of two Opia viral genotypes. Light gray bars indicate perfect nucleotide identity, and thin vertical black lines indicate SNPs. The three genes labeled in brown are present in both Opia-3708 (top) and Opia-19564 (bottom) but are absent in some other genotypes. Light blue genes are cysteine-rich (For 3708L, 10, five, nine, five, 10; for Opia-19564, three, 14, five, nine, four, 10 cysteines per protein). Dark blue genes encode ParB-like proteins that are very divergent between some genotypes (Opia-3708 vs. Opia-66823, 67881). Before the three-gene indel and after the ParB-like gene, the ∼35 kb regions of all genomes are essentially identical. (B) Phylogenetic tree including the 17 larger cysteine-rich proteins from the variable regions of seven Opia genomes (numbers represent genome names) (for context, see Supplemental Material). Proteins with identical sequences occur in five different combinations across the genotypes. (C) Comparison of the active site region of PacI (with nine cysteines) and the structure of Opia-19564 protein 16 (with 10 cysteines; silver). Active site residues of PDB 3m7k (gold) are displayed based on figure 1 of Shen et al. (2010). The critical ββα-metal motifs are well aligned. Tyrosine and histidine residues exist in proximity to the active site, although their locations differ somewhat, possibly owing to fold inaccuracy.