Figure 2.
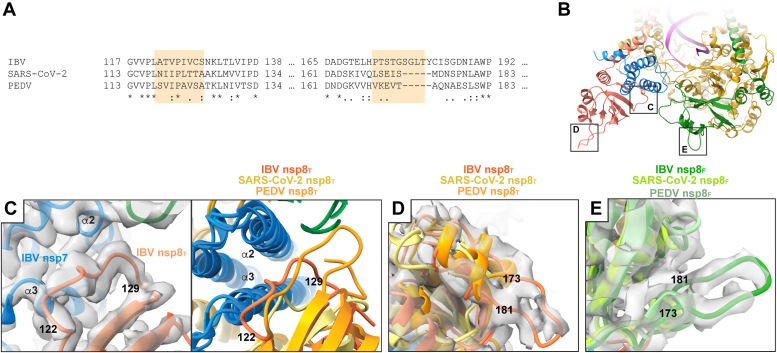
Sequence divergent features in coronavirus nsp8.A, a multiple sequence alignment of IBV, SARS-CoV-2, and PEDV nsp8 highlights sequence and structurally divergent regions. Highlighted regions correspond to the numbered IBV nsp8 loops in C–E. B, an overview of the IBV replication-transcription complex indicates structurally divergent nsp8 regions shown in greater detail in C–E. C, a focused view of IBV nsp8T 122 to 129 shows conformational heterogeneity across genera [PEDV (8URB) and SARS-CoV-2 (6XEZ)]. Reconstructed IBV RTC density is shown to support the observed conformation. D, a focused view of the nsp8T 173 to 181 loop shows an extended loop conformation (IBV RTC reconstructed density) with superimposed PEDV and SARS-CoV-2 nsp8T adopting alternate conformations. E, a focused view of nsp8F 173 to 191 showing an extended loop conformation (IBV reconstructed density) with superposed PEDV and SARS-CoV-2 nsp8F adopting alternate conformations. IBV, infectious bronchitis virus; nsp, nonstructural protein; PEDV, porcine epidemic diarrhea virus; RTC, replication–transcription complex.