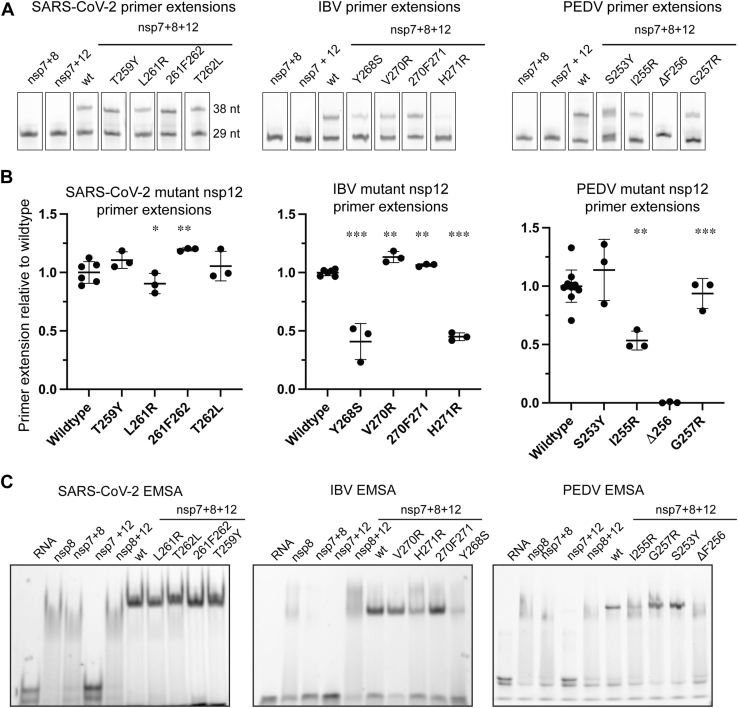
Figure 5.
Mutant nsp12 primer extensions.A, point mutations and insertions of nsp12 from SARS-CoV-2 (left), IBV (center), and PEDV (right) were tested for their ability to form the core-RTC and extend RNA in vitro. Uncropped gels are presented in Fig. S11. B, for each coronavirus, primer extension results are presented as % activity of the polymerase complex with respect to wildtype nsp12. Error bars indicate standard deviation of the technical replicates (3–12). Using an unpaired t test comparing each mutant to wildtype, “∗” denotes p < 0.05, “∗∗” denotes p < 0.001, and “∗∗∗” denotes p < 0.0001. Primer extensions were carried out in 10 mM Tris-Cl (pH8), 2 mM MgCl2, 1 mM DTT, and 10 mM NaCl (SARS-CoV-2 and PEDV) or 100 mM K-Glu (IBV). Reagents were combined at concentrations of nsp12—500 nM, nsp7—1.5 μM, nsp8—1.5 μM, and RNA—250 nM. After the addition of NTPs (500 μM), extensions ran for 1 min at room temperature (SARS-CoV-2) or 30 °C (PEDV), or 30 min at 30 °C (IBV). C, point mutations and insertions of nsp12 from SARS-CoV-2 (left), IBV (center), and PEDV (right) were further tested for their ability to form the core-RTC in vitro using an electrophoretic mobility shift assay. Complexes were assembled in buffer containing 10 mM Tris-Cl (pH 8), 2 mM MgCl2, 1 mM DTT, and 10 mM NaCl (SARS-CoV-2 and PEDV) or 10 mM potassium glutamate (IBV). Proteins were combined at nsp12—1 μM, nsp7—3 μM, and nsp8—3 μM with RNA at 250 nM. Complexes were assembled at room temperature before being analyzed via native-PAGE. Uncropped raw images of gels are presented in Fig. S10. EMSA, electrophoretic mobility shift assay; IBV, infectious bronchitis virus; nsp, nonstructural protein; PEDV, porcine epidemic diarrhea virus; RTC, replication–transcription complex.