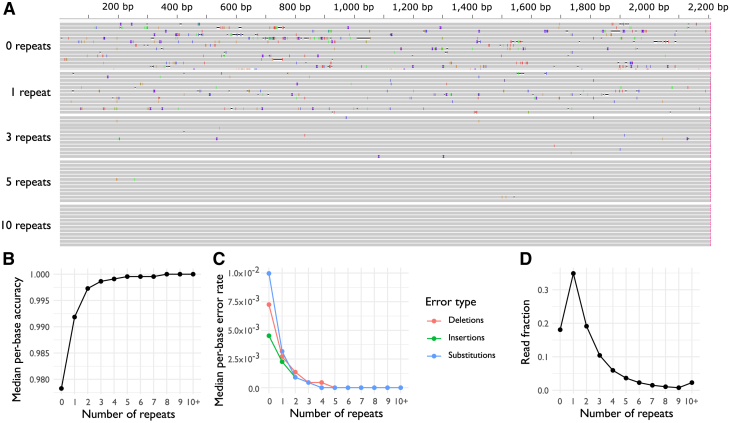
Figure 2.
Number of repeats per read influences the error rate of R2C2 sequencing
(A) Consensus reads from an AAV2 cap gene library, aligned to AAV2. Reads are separated by number of repeats used to generate the consensus sequences, where reads with zero repeats are a single instance of AAV2 cap, reads with one repeat contain two instances, and so on. Vertical purple bars indicate insertions, horizontal black lines indicate deletions, and colored bars (blue, red, orange, green) represent substitutions. (B) Median per-base accuracy for consensus reads, as a function of number of repeats. (C) Median per-base occurrences of substitutions, insertions and deletions, as a function of number of repeats. (D) Number of repeats observed in concatenated reads, expressed as a fraction of the library total.