Figure 3.
Characterization of a shuffled AAV2-N496D, AAV3b, AAV8, and AAV9 capsid library with R2C2 sequencing and AAVolve
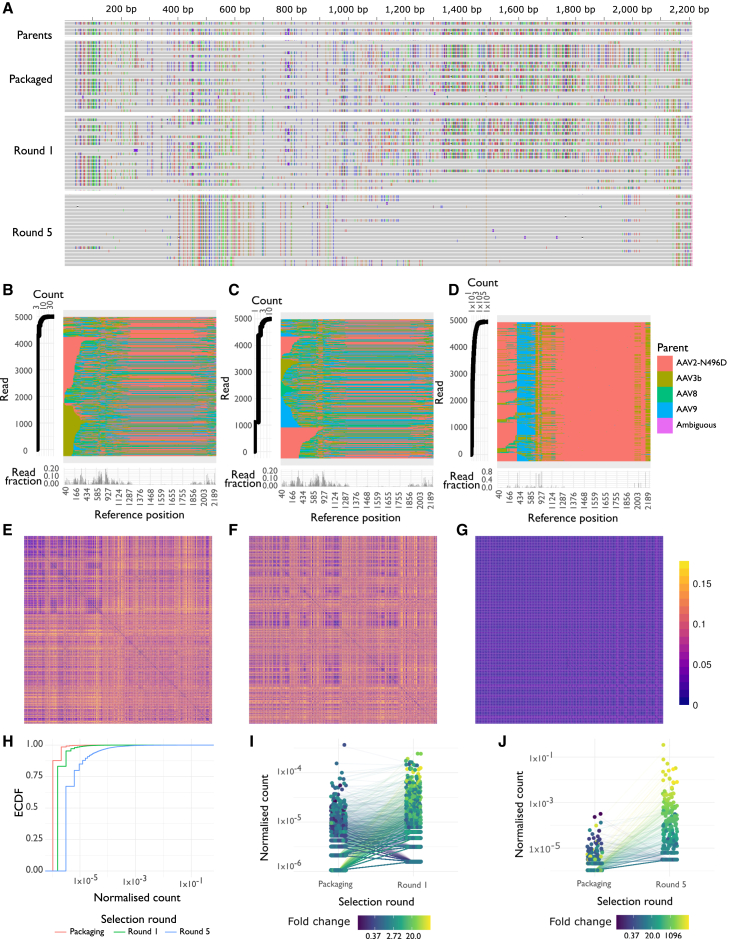
(A) Alignment of parental sequences (top) and consensus reads at three stages of selection (packaged vector, after one round and after five rounds). Variants relative to the AAV2-N496D reference are represented by colored bars (green, red, blue, orange) in each read, with variants in the library inherited from one of the four parents. (B–D) The most prevalent 5,000 unique sequences in the packaged library (B), after one round of selection (C), and after five rounds of selection (D). In (B)–(D), each row in the center represents one read, colored by most likely parent at each variant, and at left is the corresponding count for the corresponding read. (Bottom) Frequency of breakpoints occurring at each position for the whole library; that is, the fraction of reads where there are two different parents at either side of each position in the reference. (E–G) Distance matrices, reflecting fraction of aligned amino acids differing between pairs of sequences, for the most prevalent 1,000 unique sequences in each library. Distance matrices are shown for the packaged library (E), after one round of selection (F), and after five rounds of selection (G). Reads are arranged along each axis in order of prevalence in the library. (H) ECDF for the normalized counts for each of the three stages of selection: packaged vector, after one round, and after five rounds of selection. Counts were normalized by dividing the count for each unique sequence by the total library size. (I and J) Change in normalized read count for individual capsids, between the packaged library and after one round of selection (I), or between the packaged library and after five rounds (J). Each line represents an individual sequence that was observed in both sequencing libraries, colored by the fold change during selection. Counts were normalized by dividing the count for each unique sequence by the total library size.