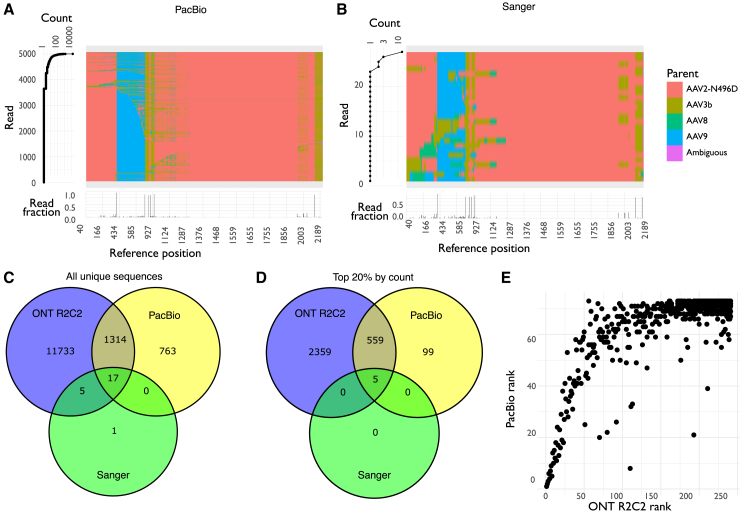
Figure 4.
PacBio HiFi and Sanger sequencing of a shuffled AAV2-N496D, AAV3b, AAV8, and AAV9 capsid library after five rounds of selection
(A and B) Overview of sequencing data for PacBio HiFi (A) or Sanger (B) sequencing, showing counts (left) and assigned parents (top right) for the top 5,000 unique sequences (A) or all clones (B), and the location and frequency of breakpoints for the whole dataset (bottom right). In (A) and (B), each row in the center represents one unique sequence, colored by most likely parent for each variant position, and at left is the corresponding count for that read. (Bottom) Frequency of breakpoints occurring at each position for the whole library; that is, the fraction of reads where there are two different parents at either side of each position in the reference. (C and D) Venn diagrams showing the number of shared sequences between the datasets for all unique sequences (C) or the unique sequences with counts in the top 20% for each dataset (D). (E) Correlation between ranks for unique sequences in the PacBio HiFi and Nanopore R2C2 datasets. Unique sequences were ranked by their count, and only the ranks for unique sequences that appeared in both datasets are shown.