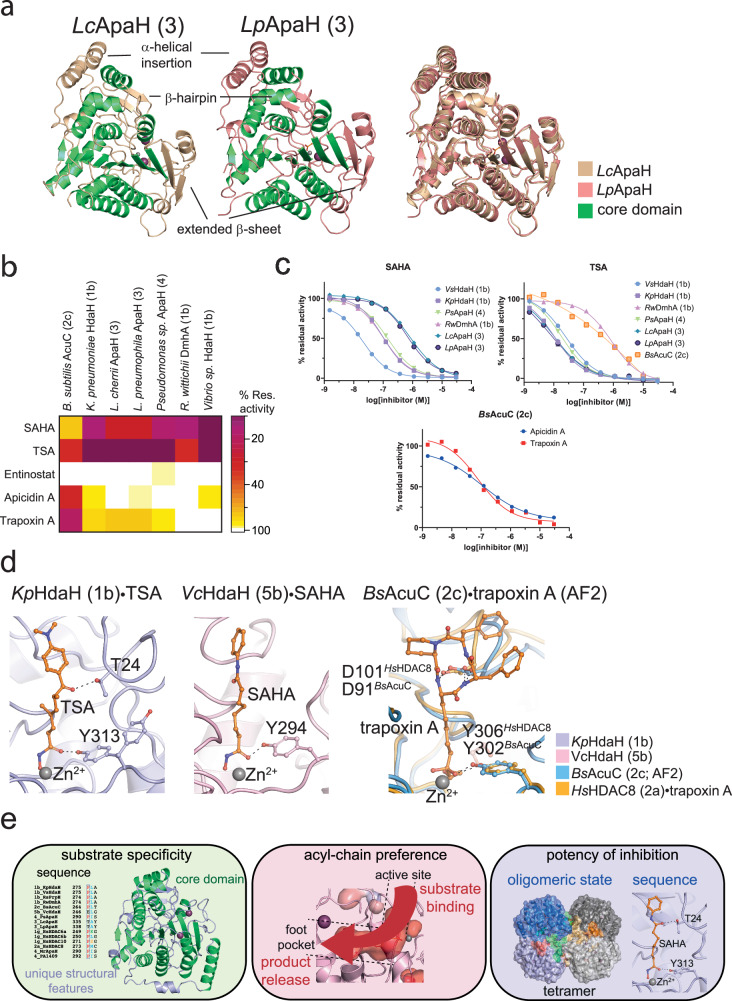
Fig. 8. Structural features of cluster 3 containing Legionella virulence factors and inhibition of bacterial deacylases by hydroxamate inhibitors and cyclic peptides.
a The cluster 3 enzymes LcApaH (3) and LpApaH (3) are bacterial virulence factors. Structurally the cluster 3 enzymes contain two distinct features. The eight-stranded parallel β-sheet is extended by two antiparallel β-strands (LcApaH (3): brown; LpApaH (3): red) and there are two additional α-helices in the catalytic core domain. b Heatmap of the pre-screening of selected bacterial deacylases by the hydroxamate inhibitors SAHA and trichostatin A (TSA), the benzamide inhibitor entinostat (MS-275), and the cyclic peptides apicidin A and trapoxin A. The color code represents the residual activity after 1 h incubation at 37 °C. The experiments were performed in two independent replicates (n = 2). Shown are the means. Source data are provided as Source Data file. c Dose-response curves obtained for inhibition of bacterial deacylases with the hydroxamate inhibitors SAHA, TSA, and the cyclic peptides apicidin A and trapoxin A. The experiments were performed in two independent replicates (n = 2). Shown are the means. Source data are provided as Source Data file. d The hydroxamate inhibitors SAHA and/or TSA use an identical inhibitory mechanism for bacterial deacylases and for mammalian HDACs. For BsAcuC (2c) an AlphaFold2 model was created to show interaction with trapoxin A by superposition with the structure of HsHDAC8 (2a)•trapoxin A (PDB: 5VI6). For binding of the cyclic inhibitors formation of hydrogen bonds between the main-chain amide of the cyclic peptides and the side chain of a conserved Asp (BsAcuC (2c): Asp91; HsHDAC2 (2a): Asp100; HsHDAC8 (2a): Asp101) is important. e Model of the molecular mechanisms underlying the observed substrate and acyl-chain type preferences of bacterial deacylases. Bacterial deacylases apply three major mechanisms for their activity and for the determination of substrate specificity. Left: Oligomerisation determines the accessibility of substrates to the active site. Middle: The architecture of the foot pocket determines acyl-chain preference. Right: Differences in the sequences and presence of additional structural features determine substrate promiscuity, acyl-chain preference, and the inhibitory potency by different classes of inhibitors.