Abstract
Non-Hodgkin lymphomas (NHL) commonly occur in immunodeficient patients, both those infected by human immunodeficiency virus (HIV) and those who have been transplanted, and are often driven by Epstein-Barr virus (EBV) with cerebral localization, raising the question of tumor immunogenicity, a critical issue for treatment responses. We investigated the immunogenomics of 68 lymphoproliferative disorders from 51 immunodeficient (34 post-transplant, 17 HIV+) and 17 immunocompetent patients. Overall, 72% were large B-cell lymphoma and 25% were primary central nervous system lymphoma, while 40% were EBV+. Tumor whole-exome and RNA sequencing, along with a bioinformatics pipeline allowed analysis of tumor mutational burden, tumor landscape and tumor microenvironment and prediction of tumor neoepitopes. Both tumor mutational burden (2.2 vs. 3.4/Mb, P=0.001) and numbers of neoepitopes (40 vs. 200, P=0.00019) were lower in EBV+ than in EBV– NHL, regardless of the immune status. In contrast both EBV and the immune status influenced the tumor mutational profile, with HNRNPF and STAT 3 mutations observed exclusively in EBV+ and immunodeficient NHL, respectively. Peripheral blood T-cell responses against tumor neoepitopes were detected in all EBV– cases but in only half of the EBV+ ones, including responses against IgH-derived MHC-class-II restricted neoepitopes. The tumor microenvironment analysis showed higher CD8 T-cell infiltrates in EBV+ versus EBV– NHL, together with a more tolerogenic profile composed of regulatory T cells, type-M2 macrophages and an increased expression of negative immune-regulators. Our results highlight that the immunogenomics of NHL in patients with immunodeficiency primarily relies on the tumor EBV status, while T-cell recognition of tumor- and IgH-specific neoepitopes is conserved in EBV– patients, offering potential opportunities for future T-cell-based immune therapies.
Introduction
Non-Hodgkin lymphomas (NHL) are highly prevalent in severely immunodeficient patients such as patients infected with human immunodeficiency virus (HIV) and transplant recipients. In both cases, the incidence is higher than in immunocompetent individuals, with a standardized incidence ratio of 11.5 for HIV-related lymphomas and 8 for post-transplant lymphoproliferative disorders (PTLD).1-3 The Epstein-Barr virus (EBV) is present in 50-70% of immunodeficient NHL and has been established as a strong oncogene.4,5 Several targeted sequencing approaches have shown that the mutational landscape of EBV+ NHL differs from that of EBV– NHL, with a lower number of mutated genes in the former.6-11 In addition, immunodeficient patients display alterations of the normally strong cellular immunity directed against viral antigens,12,13 potentially leading to immune escape.14-17 Finally, almost 10% of immunodeficient patients, the majority of whom are EBV+, have a central nervous system (CNS) localization compared to 1% in the immunocompetent population.4,5,18 As the CNS acts as an immunologically isolated site with very specific immune features, it is hypothesized that the tumor-specific and EBV-specific immune responses are even less active in this tissue, thus favoring immune escape. The remarkable successes of immunotherapies in solid tumors rely on the level of tumor immunogenicity.19-21 The tumor neoantigens that surround tumor mutations and are presented by the patients’ major histocompatibility (MHC) molecules allow the immune system to distinguish cancer from non-cancer cells and emerge as major factors for antitumor immunity, response to immunotherapies and the development of targeted treatments. Indeed, the tumor mutational burden (TMB) and the abundance of predicted immunogenic mutations are associated with higher levels of responses to immune checkpoint inhibitors and increased survival of patients22–24 while personalized vaccine approaches have successfully induced neoantigen-specific T cells associated with clinical responses.19,25-28 However limited data exist for B-cell malignancies which are very specific due to the ability of tumor cells to present tumor neoantigens with MHC class-II molecules and to produce highly mutated immunoglobulins (Ig) that may be immunogenic. Furthermore, studies in immunocompetent patients suggested that neoantigens derived from the lymphoma Ig heavy- or light-chain variable regions with a strong bias for an MHC-II presentation may be particularly critical immune targets.29,30 However, the recognition of these peculiar Ig-based neoantigens has never been analyzed in immunodeficient patients.
We hypothesized that the immunogenomics and the tumor microenvironment (TME) of NHL occurring in immunodeficient patients might be influenced by the lower immune pressure, the frequent EBV oncogenesis and the immune-privileged sites in these patients. To further investigate this hypothesis, we developed the IDeATIon project, a multicenter, prospective study of PTLD and HIV-related lymphoproliferative disorders occurring in a large series of immunodeficient patients or in immune-privileged sites such as the CNS. We took advantage of whole tumor DNA and RNA sequencing to compare the TMB, numbers of tumor neoepitopes, neoepitope-specific T-cell responses and TME as a function of both tumor EBV status and the patients’ immune status.
Methods
Patients
From 2019 to 2022, consecutive HIV-infected patients or transplant recipients with treatment-naïve lymphoproliferative disorders were enrolled in the IDeATIon project (ClinicalTrials.gov NCT03706625). Immunocompetent patients with diffuse large B-cell lymphomas (DLBCL) not otherwise specified (NOS) were included as comparators. Diagnostic tissue biopsies and blood were collected at lymphoma diagnosis and before any treatment (Online Supplementary Materials). All patients gave written informed consent (Centre de Ressources Biologiques authorization N. 18.06.46). The protocol was approved by the French national Institutional Review Board (N. 2018-A01099-46) and the “Commission Nationale de l’Informatique et des Libertés” (N. 918222) and performed in accordance with the Declaration of Helsinki.
Immunoglobulin neoepitope prediction
Using RNA-sequencing fastq files, sequences and frequencies of the Ig productive clonotypes were derived from MixCR-V3.0.31 Reads were aligned to reference V, D, J and C genes of the B-cell receptor. Each final IgH chain clonotype was identified by a CDR3 sequence. The FR3-CDR3-FR4 sequence of the dominant clonotype (≥15% of all clonotypes) was then tested for binding affinity score in NetMHC 4.032 and NetMHCIIpan 3.233 asking for 8-10 mers and 15 mers for MHC-I and -II binding, respectively. The number of neoepitopes with a score ≤500 nM was determined.
Neoepitope-specific T-cell expansion and functional validation
The most relevant non-Ig derived neoepitopes were selected upon their presentation by expressed HLA molecules or |32-microglobulin (read per kilobase per million, RPKM ≥1), and the highest pVACseq priority score for single nucleotide variations (Online Supplementary Materials), and the highest binding affinity score for indels derived from the same somatic variant. When numbers of candidate neoepitopes were >60, the stability filter (NetMHCstab) was used to select the 60 strongest binders. For Ig-derived neoepitopes, 8-14 mer peptides surrounding the mutation for MHC-I peptides and 25 mer peptides overlapping by 20-24 amino acids and spanning the complete mutated predicted sequences for MHC-II peptides were selected.
A maximum of 60 peptides corresponding to neoepitopes were synthetized per patient (GeneCust, Boynes, France). Frozen peripheral blood mononuclear cells were thawed and co-cultured with each patient’s personalized pooled peptides (2 μg/mL) in RPMI medium enriched with interleukin (IL)-2 (10 UI/mL) (Roche, Bâle, Suisse), IL-7 (25 ng/ mL) and IL-15 (25 ng/mL) (BioLegend, San Diego, CA, USA). On day 10, cell reactivity was tested in a triplicate interferon- enzyme-linked immunospot assay after stimulation with personalized pooled peptides (2 μg/mL per pool), medium alone and phytohemagglutinin as negative and positive controls, respectively. When enough cells were available, additional assays were performed with MHC-I or MHC-II pooled peptides, or each individual peptide. After stimulation for 20 hours, numbers of spot-forming cells (SFC) were read and mean triplicate SFC numbers were calculated, after background subtraction, with a positivity threshold of 50 SFC/106 cells.12,13
Gene expression and cell type abundance profiling
Tumors were digitally quantified for immune genes with a targeted gene panel including T-cell function, and positive and negative immune regulation (Online Supplementary Materials). Immune deconvolution analysis was performed with CIBERSORTx34 version, using expression matrix estimating 22 immune cell types using 547 signature genes (LM22).35 Whole exome and RNA sequencing, MHC-I and -II restricted neoepitope prediction, differential gene expression, gene ontology enrichment analysis, T-cell receptor analysis and the statistical analysis are detailed in the Online Supplementary Materials.
Results
Patients’ characteristics
Sixty-eight patients with NHL were enrolled, including 51 immunodeficient patients, who were either transplant recipients (N=34 PTLD) or HIV-positive (N=17), and 17 immunocompetent patients with DLBCL (Table 1). The median age was 57 years and 72% of patients were male. Diseases were systemic in 75% of cases and cerebral in 25% of cases. Two patients with both systemic and CNS localization were classified as having systemic disease. The most frequent subtypes of lymphoproliferative disorder were large B-cell lymphoma (LBCL) (72%: 34 cases of DLBCL NOS, 12 cases of primary CNS lymphoma, 2 cases of human herpesvirus 8-positive DLBCL NOS and 1 case of high-grade B-cell lymphoma NOS), followed by polymorphic lymphoproliferation (9%), Burkitt lymphoma and plasmablastic lymphoma (6% each). The tumors were EBV+ in 40% cases, all in the immunodeficient group (47% and 65% in PTLD and HIV+ patients, respectively). Both CNS and systemic localizations were equally distributed between EBV+ and EBV– NHL. All but one CNS NHL in immunodeficient patients were EBV+. Among the eight EBV+ tumors tested for EBV antigen expression and latency status, three tumors displayed latency I, two patients displayed latency II and three other ones displayed latency III status. No significant differences were observed between EBV+ and EBV– NHL regarding time from transplantation to NHL diagnosis (6.9 years, interquartile range [IQR], 2.85-13.11] versus 10.2 years (IQR, 8.80-20.41) (P=0.06), median CD4 counts at NHL diagnosis (266 and 222/mm3), time from diagnosis of HIV infection to NHL diagnosis (4.1 years [IQR, 2.49-21.53] versus 21.6 years [IQR, 4.92-23.94], P=0.7) and overall survival (not available and 5.3 years, P=0.21) (Online Supplementary Figure S1). The median follow-up from the diagnosis of NHL was 1.8 years.
The tumor mutational burden is lower in Epstein-Barr virus-positive than Epstein-Barr virus-negative non-Hodgkin lymphoma
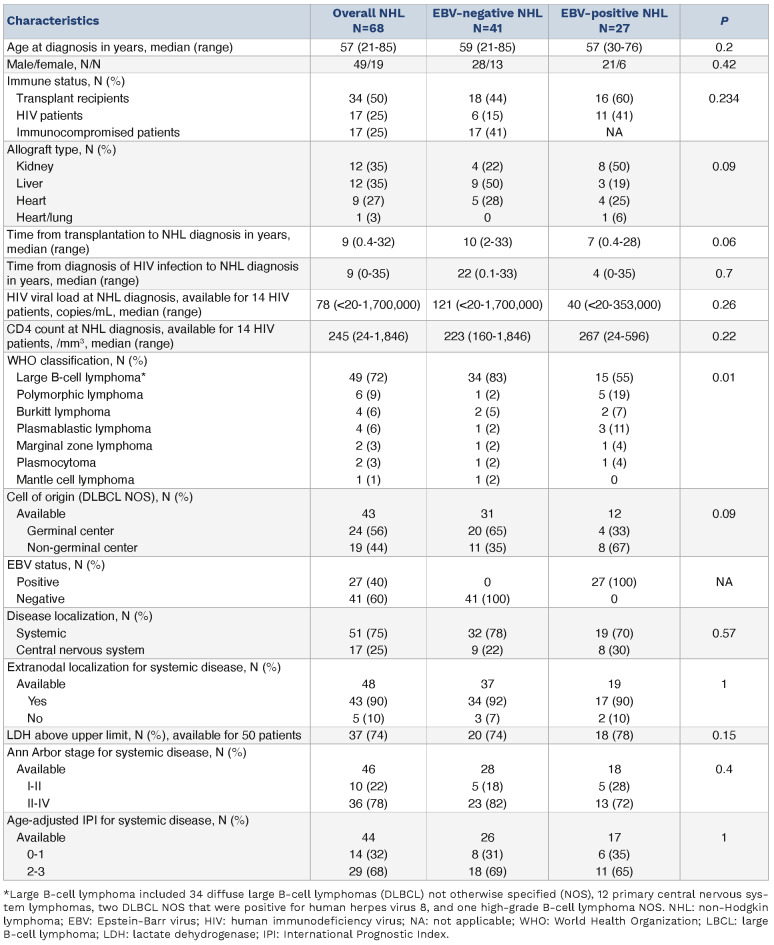
We investigated the TMB in a whole-exome sequencing (WES) analysis of paired tumor and normal samples (Online Supplementary Methods, Online Supplementary Figure S2). The median TMB was 3.01/Mb (IQR, 1.81-5.06) similar to the previously described burden for DLBCL patients (Online Supplementary Figure S3).36 However, the median TMB was lower in EBV+ NHL than in EBV– NHL, both among the whole (immunodeficient + immunocompetent) NHL population (2.2 vs. 3.4/Mb, P=0.001) and among the immunodeficient subjects only (2.2 vs. 4.6/Mb, P=0.0075) (Figure 1A). Similarly, when restricting the analysis to the predominant LBCL subgroup (N=49), a lower median TMB was observed in EBV+ than EBV– LBCL both among the whole (immunodeficient + immunocompetent) population (2.2 vs. 3.4/Mb, P=0.00071) (Figure 1A) and among the immunodeficient patients only (1.9 vs. 4.9/Mb, P=0.00078) (Online Supplementary Figure S4A). There were no differences in TMB according to immune status when comparing immunocompetent, PTLD and HIV patients both in the overall population (Figure 1B) and in the EBV– one (Online Supplementary Figure S4B). By contrast, the TMB differed slightly according to disease localization (3.2/Mb in systemic vs. 2.3/Mb in CNS disease, P=0.046) (Figure 1C). Importantly, a higher TMB was associated with a significantly longer median overall survival in EBV– patients (overall survival not reached for TMB >3/Mb vs. 2.2 years for TMB <3/Mb, P=0.01) while there was no difference in EBV+ ones (Figure 1D) (see Online Supplementary Methods for the determination of the TMB cut point).
Table 1.
Patients’ characteristics at diagnosis.
Figure 1.
The tumor mutational burden is lower in Epstein-Barr virus-positive non-Hodgkin lymphoma than in Epstein-Barr virus-negative cases. (A) Tumor mutational burden (TMB), defined as the number of mutations per megabase; log10, according to Epstein-Barr virus (EBV) status among 68 patients with non-Hodgkin lymphoma (NHL) on the left, 51 immunodeficient NHL patients in the middle, and the 49 patients with large B-cell lymphoma on the right. Red and green denote EBV-negative and -positive NHL respectively. (B) TMB according to immune status. Salmon, blue and yellow colors denote immunocompetent patients, transplant recipients and patients infected with human immunodeficiency virus, respectively. (C) TMB according to disease localization. Pink and blue denote systemic and central nervous system localization, respectively. Wilcoxon test. (D, E) Overall survival depending on TMB >3/Mb among EBV– (D) and EBV+ (E) cases. Kaplan-Meier analysis. ID: immunodeficient; LBCL: large B-cell lymphoma; CNS: central nervous system.
Epstein-Barr virus, immune status and disease localization impact the mutational profile of non-Hodgkin lymphoma
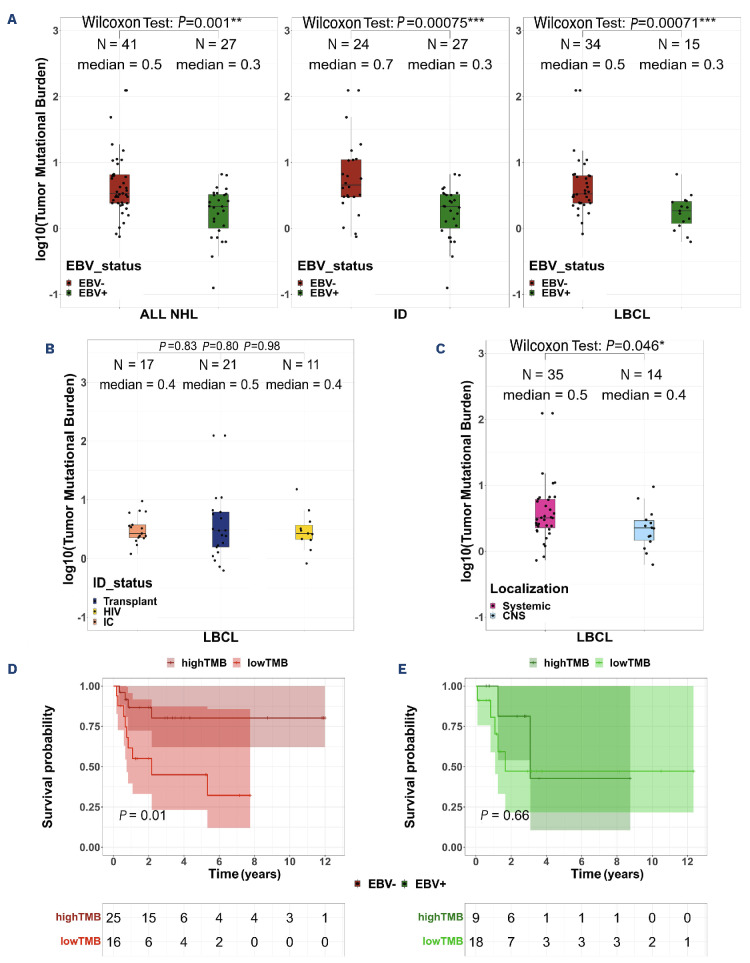
Next, we deciphered the mutational profile of these lymphomas and observed some frequently mutated genes, such as PCLO (25% cases), CSMD3 (24%), TP53 (24%), FAT4 (19%) and MYD88 (16%), as expected (Online Supplementary Figure S5A). However, when focusing on the LBCL cases we observed a dysbalance in the mutational profile depending on both the EBV and immune status of the patients. Indeed, in EBV+ LBCL, the most frequently mutated genes were TYW1 (27%) and STAT 3, HNRNPF, TNRC18, PARP10 and KDM3A (20% each), while TP53 (35%), MYD88 (32%), CD79B (24%) and FOXQ1, STAT 3, KRT3 and ZNF574 (15% each) were the most frequently mutated genes in EBV– disease (Figure 2A, Online Supplementary Table S1). Furthermore, some mutations were observed only in EBV+ NHL, such as HNRNPF (20%), or only in EBV– ones, such as MYD88 and CD79B mutations (32% and 24%, respectively). Finally, TP53 mutations were observed almost exclusively in EBV– NHL (35% vs. 7%, P=0.001). After false discovery rate (FDR) correction, no significant differences remained.
The immune status (immunocompetent or immunodeficient) co-influenced this tumor profile with TP53 present in 34% of immunodeficient LBCL (vs. 12% in immunocompetent DLBCL), all but one of which were in the EBV– group. Furthermore, STAT3, TYW1 and KDM3A mutations were found only in immunodeficient LBCL (25%, 25% and 19%, respectively), regardless of EBV status, while they were absent in immunocompetent DLBCL (Figure 2B, Online Supplementary Table S2). As expected, the disease localization also had an impact, with TP53 mutations found only in systemic diseases (37% vs. 0%, P=0.009, FDR=0.03) and MYD88 and CD79B mutations being more frequent in CNS than in systemic diseases (50% vs. 11%, P=0.006, FDR=0.03, and 43% vs. 6%, P=0.04, FDR=0.03, respectively) (Online Supplementary Figure S5B). Finally, the six polymorphic lymphoproliferations (5 EBV+ and 1 EBV–) showed a distinct mutational profile with PCLO as the most recurrently mutated gene (Online Supplementary Figure S5C). Within the four cases of Burkitt lymphoma, the two genes recurrently mutated were MYC and IGLL5 (Online Supplementary Figure S5D). In addition, we identified significant recurrent somatic copy number alterations (SCNA) in the WES data (Online Supplementary Methods). When focusing on the 47 LBCL, we successfully identified a total of 13 arm-level and 12 focal regions displaying copy gain, along with five arm-level and six focal regions with copy loss (arm q-value ≤0.1, focal regions q-value ≤0.25) (Online Supplementary Figure S6A). The observed frequencies of these SCNA ranged from 2% to 55%. In immunocompetent DLBCL, SCNA encompassed amplifications of 1q, 18q, 21q, 7q22, 8q24, and 19q13, and deletions involving 6q, 17p, 1p36, and 1p13, consistently with prior reports.37 Additionally, we observed SCNA in genes previously reported in DLBCL,38 including PIK3CA (25%), PRDM1 (38%), MYC (19%), CDKN2A (21%), PTEN (14%), ETV6 (42%), STAT6 (42%), B2M (23%), and TP53 (42%) (Online Supplementary Table S3). Furthermore, we uncovered previously unreported amplifications of chromosome 16 in 34% of the LBCL patients in our dataset. Notably, factors influencing those SCNA included the EBV status with focal amplification peaks, in EBV+ cases, of the 9q34.13, a region encompassing the NOTCH1 and JAK2 genes (Online Supplementary Figure S6B). The immune status also influenced SCNA with more significantly frequent amplification at 1q22 (33%; q-value=0.0185/29%; q-value >0.25), 8q24.3 (26%; q-value=0.04/17% q-value >0.25), 9q34.3 (47%; q-value=0.0008/24%; q-value >0.25), 11q13.1 (43%; q-value=0.0032/23%; q-value >0.25), 17q25.3 (36%; q-value=0.0219/29%; q-value >0.25), 22q13.31 (33%; q-value=0.1577/5% q-value >0.25) genomic regions in the immunodeficient LBCL versus immunocompetent DLBCL groups. Additionally, some deletions were prevalent at 1p36.32 (23%; q-value=0.0361/13%; q-value >0.25) and 15q12 (36%; q-value=0.0018/29% q-value >0.25) (Online Supplementary Figure S6C), affecting genes such as IL10, MYC, CD45, IRF8, IL17RA, and CDKN2C, among others. Systemic localizations also played a role with more frequent deletions affecting the human leukocyte antigen (HLA) locus (6p21.31: 9%; q-value=0.22/0%; q-value >0.25), BCL6 (3q29: 19%;q-value=0.19/0%; q-value >0.25) and IG genes (15q12: 37%; q-value=0.005/28%; q-value >0.25), together with amplifications of the 8q24 locus (33%; q-value=0.096/28%; q-value >0.25) (Online Supplementary Figure S6D).
Figure 2.
The mutational landscape of non-Hodgkin lymphoma differs by Epstein-Barr virus and immune status. (A) Co-oncoplot of the most recurrently mutated genes (≥15%) within the 49 samples of large B-cell lymphoma (LBCL) according to Epstein-Barr virus (EBV) status (EBV– on the left, EBV+ on the right). TP53, MYD88 and HNRNFP were differently mutated between the two groups but these results lost their significant value after false discovery rate (FDR) correction was applied. (B) Co-oncoplot of the most recurrently mutated genes (≥15%) within the 49 LBCL samples according to immune status (immunocompetent on the left, immunodeficient on the right). STAT 3, TYW1 and MYD88 were differently mutated between the two groups but these results lost their significant value after FDR correction was applied. Fisher exact test was used to compare categorical data. CNS: central nervous system; PTLD: post-transplant lymphoproliferative disorder; DLBCL: diffuse large B-cell lymphoma; ID: immunodeficient; IC: immunocompetent; HIV: human immunodeficiency virus.
The Epstein-Barr virus status influences the tumor immunogenicity dominated by MHC class-II restricted neoepitopes
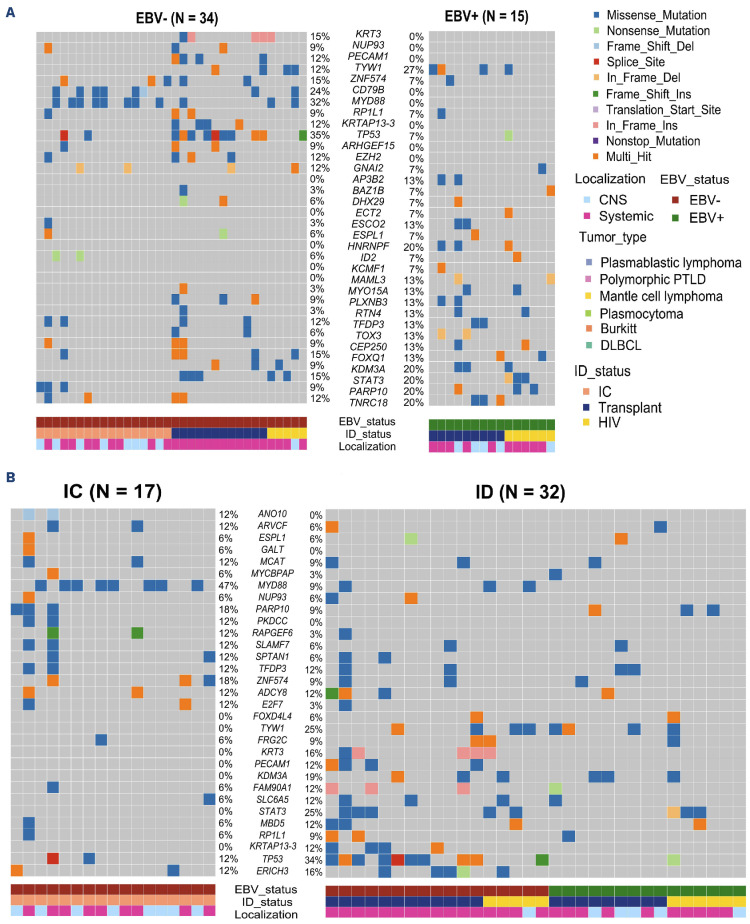
We first predicted the neoepitopes derived from non-Ig tumor variants by including parameters from the RNA sequencing. In the 30 tumors with available RNA-sequencing data (Online Supplementary Methods; Online Supplementary Figure S2B), we found a median of 149 neoepitopes per tumor, with lower numbers in EBV+ compared to EBV– NHL (40 vs. 200, respectively, P=0.00016), independently of the immune status (Figure 3A). These values were not influenced by the expression levels of the MHC molecules and B2M that did not differ between EBV+ and EBV– NHL, immunodeficient and immunocompetent patients and systemic versus CNS localization (data not shown). Furthermore, very few downregulations (RPKM <1) of HLA class I and class II were observed (3% and 9% of tumors, respectively) and no B2M downregulation or mutations were observed despite the presence of heterozygote deletions in four tested patients. Most neoepitopes were predicted to be presented by MHC class-II molecules with a median class-II/ class-I MCH ratio of 4, independently of EBV status. As RNA-sequencing data were not available for more than half the tumors, we conducted the same analysis solely based on WES. Similarly, we observed lower numbers of neoepitopes in EBV+ compared to EBV– tumors (159 vs. 501, P=0.0024) (Figure 3B), with a highly significant positive correlation between WES and RNA-based results (r=0.93, P<0.001) (Figure 3C). Importantly, the neoepitope numbers were positively correlated to the TMB (r=0.8, P<0.001) (Figure 3C). Furthermore clonal-derived neoepitopes accounted for 89% of the entire WES predicted neoepitopes with significantly higher neoepitope numbers originating from clonal compared to subclonal populations (median per patient: 301 and 32, respectively, P=0.02) (Online Supplementary Figure S7). We then analyzed the neoepitopes derived from Ig variable region genes by using the available RNA-sequencing data from the FR3-CDR3-FR4 region. A dominant tumor Ig clonotype (≥15%) was found in 24 NHL patients, with frequencies ranging from 15% to 100% (median 81%). A median of 15 neoepitopes derived from the tumor heavy chain variable region genes was found, independently of the EBV status (15 and 18 for EBV+ and EBV– samples, respectively) or immune status (15 and 15 for immunodeficient and immunocompetent samples, respectively) (Figure 3D). These Ig-derived neoepitopes were also predicted to be more frequently presented by MHC class II, with an MHC class-II/class-I ratio of 1.5.
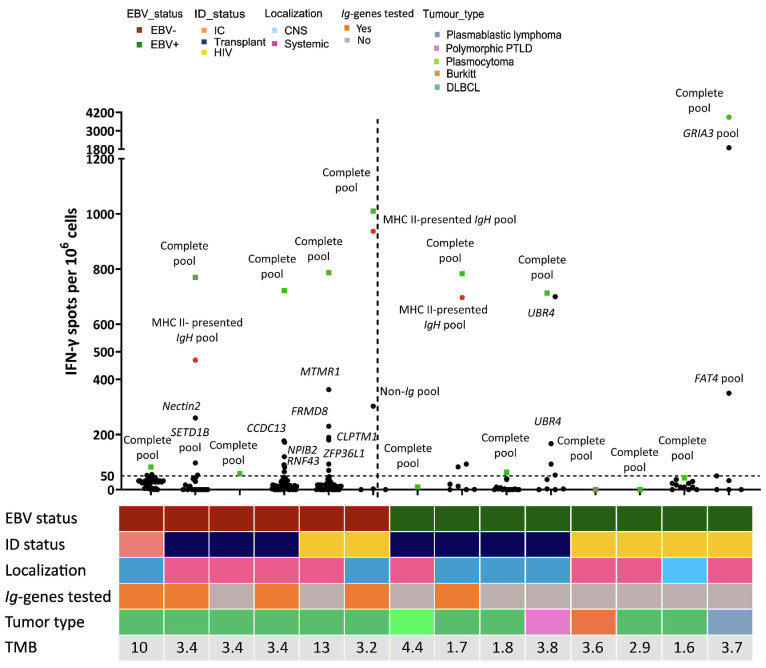
Circulating T cells specific for tumor neoepitopes, including Ig-derived neoepitopes, were detected in 71% of non-Hodgkin lymphomas
To confirm that the predicted NHL neoepitopes could drive effective antitumor immunity in immunodeficient patients, we analyzed the peripheral blood T-cell responses against the corresponding peptides in 14 samples (13 immunodeficient and 1 immunocompetent patient). We tested a similar number of variant-derived peptides from EBV+ and EBV- tumors with a median of 48 peptides per patient. Neoepitope-specific responses were detectable in ten out of the 14 (71%) patients’ samples tested, among whom 93% were immunodeficient and 50% had CNS disease. These neoepitope-specific responses were more frequently detectable, though non-significantly, in the EBV– patients, as observed among all six (5 immunodeficient + 1 immunocompetent, 100%) tested cases, than in EBV+ ones as observed in only four of the eight (50%) immunodeficient cases tested (P=0.08). The level of T-cell responses to tumoral neoepitopes did not change with expression of the EBV antigen (EBNA-2, LMP1 and EBER). The median magnitude of positive T-cell responses was 746 SFC/106 cells per patient’s peptide pool, and above 500 SFC/106 cells in the seven immunodeficient patients’ samples (Figure 4, Online Supplementary Figure S8). The magnitudes above SFC >500/106 cells were associated with a trend towards longer, though non-significantly different, time from transplantation or HIV diagnosis to NHL diagnosis and higher CD4 count in HIV patients (12 vs. 8 years, 27 vs. 9 years, 135 vs. 1,087/mm3, respectively, P>0.4). The Ig-derived neoepitopes were tested in five cases with a median number of 22 tested neoepitopes per patient (16 MHC-II and 8 MHC-I restricted) and were recognized in three cases with a 1.5- to 3-fold higher magnitude (median 697 SFC/106 cells) than against autologous non-Ig neoepitopes.
Figure 3.
The number of neoepitopes is lower in Epstein-Barr virus-positive non-Hodgkin lymphoma than in Epstein-Barr virus-negative cases. (A) Number of predicted neoepitopes (log10) from the non-Ig variants within the 31 RNA samples, according to Epstein-Barr virus (EBV) status on the left and immune status (in EBV– non-Hodgkin lymphoma) on the right. Wilcoxon test. (B) Number of predicted neoepitopes (log10) from the non-Ig variants within the 66 whole-exome sequencing samples (2 samples were excluded because of absence of germline assessment) according to EBV status. Wilcoxon test. (C) Correlation study between the number of predicted neoepitopes from the RNA and the whole-exome sequencing data (top) and between the number of predicted neoepitopes from the RNA data and the tumor mutational burden (bottom). Spearman correlation. (D) Number of predicted neoepitopes (log10) from Ig variants within the 24 RNA samples (7 samples were excluded because of dominant IgH clone <15%). Wilcoxon test. NHL: non-Hodgkin lymphoma; ID: immunodeficient; IC: immunocompetent; HIV: human immunodeficiency virus; WES: whole-exome sequencing.
Figure 4.
Neoepitope-specific T cells were detected in 71% cases, including responses directed against Ig-derived neoepitopes. Number of interferon (IFN)-γ spots (/106 cells) after peptide stimulation for the 14 tested patients (represented on the x axis). Green squares denote responses directed against the complete pool, red circles denote responses directed against Ig-derived neoepitopes and black circles denote responses directed against individual non-Ig neoepitopes. Thawed peripheral blood mononuclear cells were co-cultured with personalized pooled peptides for 10 days and then tested for reactivity using IFN-γ enzyme-linked immunospot (ELISPOT) assays. Patients were all tested for their personalized pooled peptides (named “complete pool”) and eventually for each individual peptide if the numbers of cells were adequate (named as the mutated gene). The mean numbers of spot-forming cells (SFC) from triplicate assays were normalized to the number of IFN-γ spots detected per 1x106 peripheral blood mononuclear cells after background subtraction. The threshold for ELISPOT-IFN-γ positivity was 50 SFC/106 cells. EBV: Epstein-Barr virus; ID: immunodeficiency; IC: immunocompetent; HIV: human immunodeficiency virus; CNS: central nervous system; PTLD: post-transplant lymphoproliferative disorder; DLBCL: diffuse large B-cell lymphoma; TMB: tumor mutational burden.
All recognized Ig epitopes were localized in the heavy chain region and predicted to be MHC-class II restricted. Finally, there were no neoepitopes shared between tumors, although some were derived from frequently mutated genes (in ≥10% NHL) such as FAT4, SETD1B, ZFP36L1, HMCN1, MTMR1, UBR4, KLF2, RBMX1, ALMS1 and C2ORF49. These tumor-derived neoepitopes were not found in the public immune epitope database for T-cell epitopes from pathogens and autoimmunity.
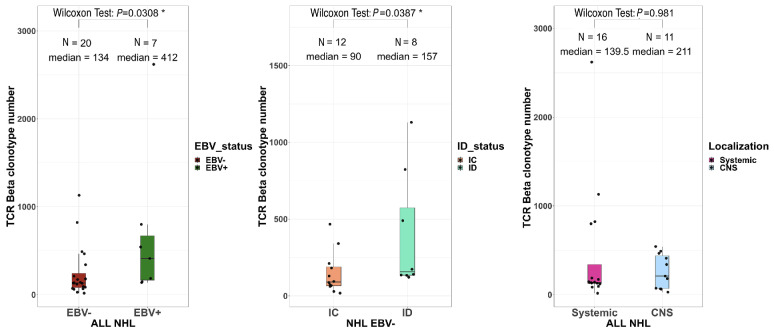
Epstein-Barr virus drives the tumor microenvironment
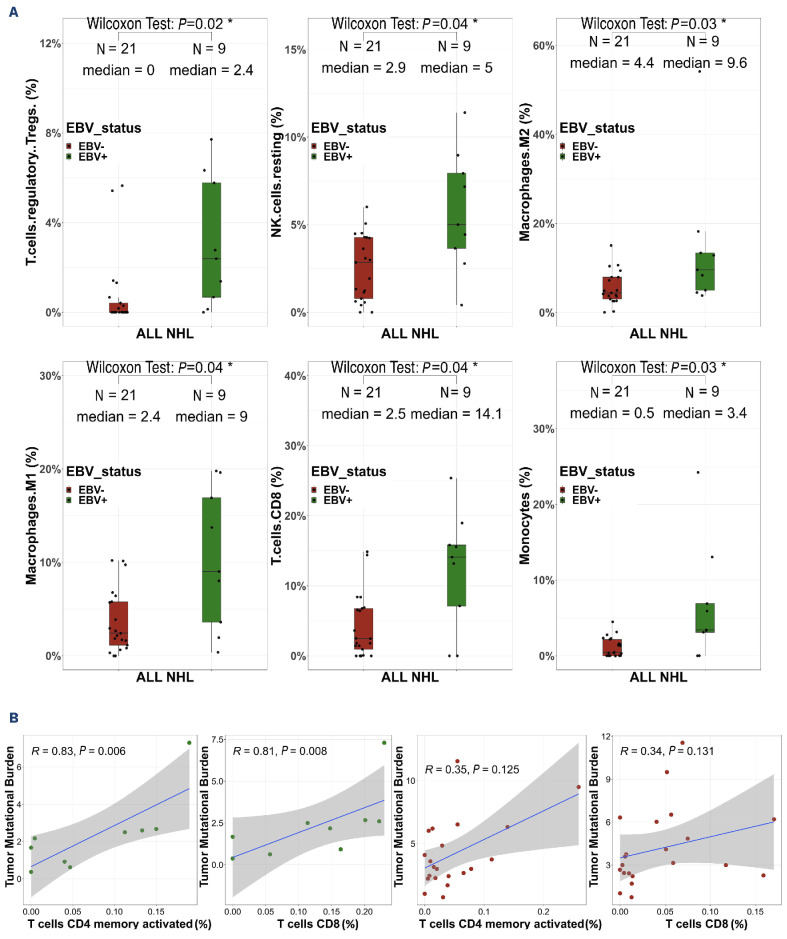
These immunogenicity results prompted us to analyze the intra-tumoral T-cell receptor (TCR) repertoire in RNA-sequencing data of 27 patients’ samples with sufficient numbers of TCR|3 reads (≥100). The EBV+ NHL contained higher numbers of unique productive clonotypes than the EBV- ones (412 vs. 134, P=0.003) (Figure 5). These numbers were also higher among immunodeficient NHL than immunocompetent ones, both in the EBV- NHL (157 vs. 90, P=0.04) and in the overall population (Online Supplementary Figure S9). In contrast, the repertoire diversity was similar between the NHL subgroups (Online Supplementary Figure S10) without any shared dominant clonotypes between patients. Finally, the TCR sequences with clone frequencies >10%, suggestive of tumor-neoantigen selection, did not show any evidence of known antigen specificity using the VDJ database. We then analyzed the TME immune cell composition to determine whether such higher TCR abundance in immunodeficient NHL was associated with selected T-cell populations (Online Supplementary Figure S11). The proportions of CD8+ T cells, regulatory T cells, resting NK cells, type-M1 and M2 macrophages and monocytes were significantly higher in EBV+ NHL than in EBV– NHL in the whole cohort (immunocompetent + immunodeficient) and tended to be higher in the immunodeficient cohort only, but were not influenced by the immune status or disease localization (Figure 6A, Online Supplementary Figures S12-S14). Conversely, the proportion of B cells was decreased in EBV+ NHL compared to EBV– NHL (14 vs. 52%, P=0.000008) (data not shown). In EBV+ NHL, there was a positive correlation between the proportions of CD8 T cells and of activated memory CD4 T cells, and the TMB (r=0.79, P=0.048; r=0.96, P=0.024, respectively) (Figure 6B). A similar trend of association was observed among EBV– cases, although not statistically significant.
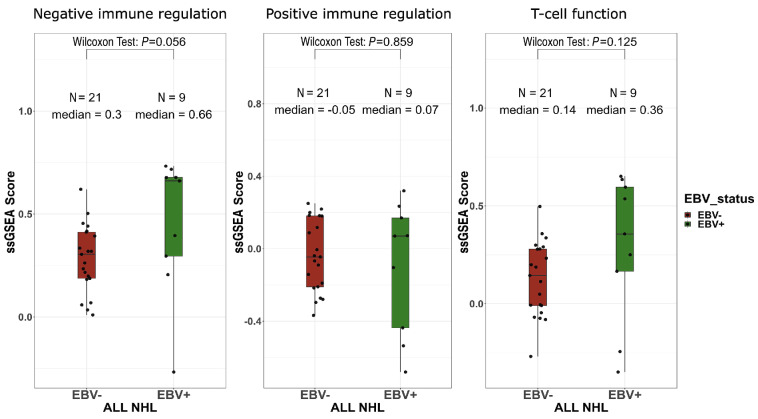
Finally, we studied the expression of a selected customized panel of relevant genes involved in intratumoral immune responses. Whereas there was no difference for genes involved in positive immune regulation or T-cell lymphocyte function, a higher non-significant expression of genes involved in negative immune regulation was observed in EBV+ NHL samples (Figure 7).
Discussion
Our comprehensive large study of the immunogenomics of NHL occurring in immunodeficient patients revealed that the presence of EBV influences both the tumor mutational burdens and profiles, along with the immune status, but also influences the tumor variant immunogenicity and the microenvironment. Although a limitation of our study may be the size of our series of NHL, particularly in the context of RNA-sequencing analysis (30 samples) for MET studies and enzyme-linked immunospot analysis (14 samples) for neoepitope-specific T-cell response studies, it is the largest series so far to be studied at the unbiased immunogenomics level. In addition, some heterogeneity in the lymphoproliferative disorders studied might have confounded some analyses given that the major lymphoma subgroup of LBCL and the systemic localization constituted respectively only 72% (N=49) and 75% (N=51) of the entire cohort. The median onset of EBV+ PTLD among transplant recipients seemed to be later than usually described and may reflect the evolving use of immunosuppressive regimens and the aging of the patients.5,39-41
Figure 5.
The intra-tumoral T-cell receptor repertoire diversity does not differ between Epstein-Barr virus-positive and -negative non-Hodgkin lymphomas. The number of unique productive T-cell receptor-β clonotypes according to Epstein-Barr virus status (left), immune status (middle) and disease localization (right). Wilcoxon test. TCR: T-cell receptor; EBV: Epstein-Barr virus; NHL: non-Hodgkin lymphoma; immunodeficient; IC: immunocompetent; CNS: central nervous system.
Figure 6.
Epstein-Barr virus drives the tumor microenvironment in non-Hodgkin lymphoma in immunosuppressed and immunocompetent patients. (A) Cell type abundance assessed with CIBERSORTx, according to Epstein-Barr virus (EBV) status. Wilcoxon test. (B) Correlation study between tumor mutational burden and memory resting CD4 T cells and CD8 T cells within EBV+ non-Hodgkin lymphoma (green circles, upper) and EBV– non-Hodgkin lymphoma (red circles, lower). Spearman correlation. NHL: non-Hodgkin lymphoma; NK: natural killer.
Our unbiased WES-based analysis allowed us to demonstrate a lower TMB in EBV+ NHL of immunodeficient patients compared to EBV– ones, confirming previously suggested data obtained by targeted approaches.6-8,42 Our data further indicate that the strong viral signaling present in EBV+ NHL reduces the need for other driver tumor mutations in lymphomagenesis, in accordance with the concept of EBV alone acting as a strong oncogene in infected B cells, especially in the context of low immune pressure.5 In line with this hypothesis, we showed that the numbers of tumor neoepitopes determined by these mutations were also lower in EBV+ NHL, suggesting that each additional mutation increases the probability of generating a significant neoantigen, thereby contributing to the overall tumor immunogenicity.
Additionally, the presence of EBV also drives specific mutational profiles. The finding that HNRNPF was significantly mutated only in EBV+ NHL might be in accordance with data showing that other ribonucleoproteins from the same HNRNPK family bind to the EBV nuclear antigen-2 (EBNA-2) and enhance viral LMP2A antigen expression.43 We also confirmed previous targeted approaches reporting the almost complete lack of MYD88, CD79B or TP53 mutations in EBV+ NHL. More importantly, our WES approach enabled us to show that an immunodeficient status was associated with STAT 3 and TYW1 mutations regardless of EBV status, with S TAT 3 being mutated only among immunodeficient patients, both HIV+ and transplant recipients. The STAT3 protein is involved in a key signaling pathway modulating multiple physiological processes and inflammatory responses, especially in the B-cell lineage. STAT 3 mutations, which are rare in immunocompetent EBV– DLBCL had been reported in half EBV* diseases, such as plasmablastic lymphoma or polymorphic lymphoproliferations, particularly in HIV+ patients.44-47 However, data for STAT 3 mutations in PTLD patients are limited.48 Our findings suggest for the first time that chronic immune stimulation and/or inflammation in these two immunodeficient settings associated with permanent antigenic stimuli (allogeneic transplant or HIV) might favor such S TAT 3 mutations, thus highlighting the potential of inhibitors of JAK-STAT signaling as a promising treatment option. In addition, STAT 3 mutations have been reported very recently to be linked with a “hot” microenvironment in PCNSL.49
The prediction and validation of the most relevant neoepitopes were allowed by our development of a robust bioinformatic method based on tumor DNA and RNA sequencing, along with in silico algorithms and inferred rules for tumor neoepitope immunogenicity. In order to consider all steps of the antigen processing required for successful neoepitope selection,50 we included factors in our pipeline that are involved in the presentation machinery, such as the HLA molecules and B2M expression, in addition to key parameters for effective anti-tumor immune responses.20,51,52 The strong correlation observed between neoepitope numbers selected from WES and RNA sequencing should facilitate future routine use. In addition to these non-viral neoepitopes, we showed, as previously suggested in immunocompetent patients,29,30 that NHL Ig-derived neoantigens contain immunodominant neoantigens, mostly presented by MHC-class II, in these immunodeficient patients. Finally, as EBNA-2, the immunodominant EBV antigen was expressed in only three of the eight EBV+ tumors evaluated, and as this or the latency state I or II status did not change the neoepitope-specific immune response in the tumors, these data, although limited, suggest that EBV antigen expression does not modify tumor neoepitope-specific immune responses. Nevertheless, it will be essential in the future to characterize both the anti-EBV and anti-neoepitope immune responses in order to assess the burden of EBV better.
The higher T-cell infiltrate observed in the TME analysis of EBV+ NHL might be in accordance with strong EBV immunodominance, regardless of immune status and without evidence of enrichment in specific TCR clonotypes. This T-cell microenvironment was composed of both CD8 and activated CD4s. In addition, a tolerogenic profile, composed of regulatory T cells, type M2 macrophages and a higher expression of negative immune regulation molecules, tended to predominate in EBV+ NHL, thus confirming that EBV might promote a more tolerogenic TME.16,17 In conclusion, our exhaustive analysis of the immunogenomic characteristics of NHL occurring in immunodeficient patients shows the major influence of EBV on tumor mutational burden and profile and on tumor neo-antigenicity. Despite the lack of frequent or public NHL driver mutations and neoepitopes preventing the development of shared immune strategies as in other cancers,26,27 the existence of T-cell responses in these immunodeficient contexts, directed against non-viral tumor neoepitopes, and particularly against IgH ones in immunodeficient patients, could pave the way for the development of future Ig-based immune therapies.
Figure 7.
Gene expression profiling tends to differ between Epstein-Barr virus-positive and -negative non-Hodgkin lymphoma. Single sample gene set enrichment analysis (ssGSEA) scores of negative immune stimulation (left panel), positive immune stimulation (middle panel) and T-cell function (right panel). ssGSEA calculates separate enrichment scores for each pairing of a sample and gene set. Each ssGSEA enrichment score represents the degree to which the genes in a particular gene set are coordinately up- or down-regulated within a sample. Wilcoxon test. EBV: Epstein-Barr virus; NHL: non-Hodgkin lymphoma.
Supplementary Material
Acknowledgments
We acknowledge all the patients included in the study. We are deeply grateful to MSDAvenir and Fondation pour la Recherche Médicale for their financial support. We thank the Centre de Ressources Biologiques (CRB) of Paris-Saclay and J. Tisserand, the CRB-C SU and Rihab Jrad, the Plateforme de Ressources Biologiques) of Hôpital Mondor, and Dr C. Barau, Dr. A. Jebanesan, and Dr. MC. Morcelet for giving and transferring tumor samples. We thank the Onconeurotek Biobank of AP-HP Sorbonne Université and Amel Dridi-Aloulou for transferring brain samples. We thank Elodie Courret for support with biological samples.
Funding Statement
Funding: The IDeaTIon project is financially supported by the MSDAvenir endowment fund (grant number DS-2017-0018). This work was in part supported by the grant INCa-DGOS-Inserm_12560 of the SiRIC CURAMUS (Cancer United Research Associating Medicine, University and Society integrated cancer research program), and by the Fondation pour la Recherche Médicale, grant number FDT202106013113 to MB.
Data-sharing statement
Data are available upon reasonable request. The French data privacy authority (CNIL, Commission Nationale de l’Informatique et des Libertés) and European regulations (GDPR, General Data Protection Regulation) do not provide for transmission of the database, nor do the information and consent documents signed by the patients. Consultation by the editorial board or interested researchers of individual participant data that underlie the results reported in the article after deidentification may nevertheless be considered, subject to prior determination of the terms and conditions of such consultation and in respect of compliance with the applicable regulations. For all inquiries, please contact drc-secretariat-promotion@aphp.fr. The R code (version 4.2.1) utilized in our research is accessible upon reasonable request. Please contact the author for correspondence to obtain access.
References
- 1.Engels EA. Cancer in solid organ transplant recipients: there is still much to learn and do. Am J Transplant. 2017;17(8):1967-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hernández-Ramírez RU, Shiels MS, Dubrow R, Engels EA. Cancer risk in HIV-infected people in the USA from 1996 to 2012: a population-based, registry-linkage study. Lancet HIV. 2017;4(11):e495-e504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blosser CD, Haber G, Engels EA. Changes in cancer incidence and outcomes among kidney transplant recipients in the United States over a thirty-year period. Kidney Int. 2021;99(6):1430-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carbone A, Vaccher E, Gloghini A. Hematologic cancers in individuals infected by HIV. Blood. 2022;139(7):995-1012. [DOI] [PubMed] [Google Scholar]
- 5.Dharnidharka VR, Webster AC, Martinez OM, Preiksaitis JK, Leblond V, Choquet S. Post-transplant lymphoproliferative disorders. Nat Rev Dis Primers. 2016;2(1):15088. [DOI] [PubMed] [Google Scholar]
- 6.Finalet Ferreiro J, Morscio J, Dierickx D, et al. EBV-positive and EBV-negative posttransplant diffuse large B cell lymphomas have distinct genomic and transcriptomic features: genetics of PTLD. Am J Transplant. 2016;16(2):414-425. [DOI] [PubMed] [Google Scholar]
- 7.Gandhi MK, Hoang T, Law SC, et al. EBV-associated primary CNS lymphoma occurring after immunosuppression is a distinct immunobiological entity. Blood. 2021;137(11):1468-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menter T, Juskevicius D, Alikian M, et al. Mutational landscape of B-cell post-transplant lymphoproliferative disorders. Br J Haematol. 2017;178(1):48-56. [DOI] [PubMed] [Google Scholar]
- 9.Yoon H, Park S, Ju H, et al. Integrated copy number and gene expression profiling analysis of Epstein-Barr virus-positive diffuse large B-cell lymphoma. Genes Chromosomes Cancer. 2015;54(6):383-396. [DOI] [PubMed] [Google Scholar]
- 10.Kaulen LD, Denisova E, Hinz F, et al. Integrated genetic analyses of immunodeficiency-associated Epstein-Barr virus- (EBV) positive primary CNS lymphomas. Acta Neuropathol. 2023;146(3):499-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guney E, Lucas C-HG, Singh K, et al. Molecular profiling identifies at least 3 distinct types of posttransplant lymphoproliferative disorder involving the CNS. Blood Adv. 2023;7(13):3307-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakid-Cordero C, Arzouk N, Gauthier N, et al. Skewed T cell responses to Epstein-Barr virus in long-term asymptomatic kidney transplant recipients. PLoS One. 2019;14(10):e0224211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakid-Cordero C, Choquet S, Gauthier N, et al. Distinct immunopathological mechanisms of EBV-positive and EBV-negative posttransplant lymphoproliferative disorders. Am J Transplant. 2021;21(8):2846-2863. [DOI] [PubMed] [Google Scholar]
- 14.Green MR, Rodig S, Juszczynski P, et al. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and post-transplant lymphoproliferative disorders: implications for targeted therapy. Clin Cancer Res. 2012;18(6):1611-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicolae A, Pittaluga S, Abdullah S, et al. EBV-positive large B-cell lymphomas in young patients: a nodal lymphoma with evidence for a tolerogenic immune environment. Blood. 2015;126(7):863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keane C, Tobin J, Gunawardana J, et al. The tumour microenvironment is immuno-tolerogenic and a principal determinant of patient outcome in EBV-positive diffuse large B-cell lymphoma. Eur J Haematol. 2019;103(3):200-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keane C, Gould C, Jones K, et al. The T-cell receptor repertoire influences the tumor microenvironment and is associated with survival in aggressive B-cell lymphoma. Clin Cancer Res. 2017;23(7):1820-1828. [DOI] [PubMed] [Google Scholar]
- 18.Dierickx D, Habermann TM. Post-transplantation lymphoproliferative disorders in adults. N Engl J Med. 2018;378(6):549-562. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Goedegebuure SP, Gillanders WE. Preclinical and clinical development of neoantigen vaccines. Ann Oncol. 2017;28(Suppl_12):xii11-xii17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69-74. [DOI] [PubMed] [Google Scholar]
- 21.Yarchoan M, Johnson BA, Lutz ER, Laheru DA, Jaffee EM. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer. 2017;17(4):209-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown SD, Warren RL, Gibb EA, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014;24(5):743-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizvi H, Sanchez-Vega F, La K, et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol. 2018;36(7):633-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leidner R, Sanjuan Silva N, Huang H, et al. Neoantigen T-cell receptor gene therapy in pancreatic cancer. N Engl J Med. 2022;386(22):2112-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li F, Deng L, Jackson KR, et al. Neoantigen vaccination induces clinical and immunologic responses in non-small cell lung cancer patients harboring EGFR mutations. J Immunother Cancer. 2021;9(7):e002531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Platten M, Bunse L, Wick A, et al. A vaccine targeting mutant IDH1 in newly diagnosed glioma. Nature. 2021;592(7854):463-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sahin U, Derhovanessian E, Miller M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547(7662):222-226. [DOI] [PubMed] [Google Scholar]
- 29.Khodadoust MS, Olsson N, Wagar LE, et al. Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature. 2017;543(7647):723-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khodadoust MS, Olsson N, Chen B, et al. B-cell lymphomas present immunoglobulin neoantigens. Blood. 2019;133(8):878-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolotin DA, Poslavsky S, Mitrophanov I, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12(5):380-381. [DOI] [PubMed] [Google Scholar]
- 32.Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics. 2016;32(4):511-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen KK, Andreatta M, Marcatili P, et al. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology. 2018;154(3):394-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steen CB, Liu CL, Alizadeh AA, Newman AM. Profiling cell type abundance and expression in bulk tissues with CIBERSORTx. Methods Mol Biol. 2020;2117:135-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Australian Pancreatic Cancer Genome Initiative ICGC Breast Cancer Consortium ICGC MMML-Seq Consortium et.al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24(5):679-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitz R, Wright GW, Huang DW, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378(15):1396-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luskin MR, Heil DS, Tan KS, et al. The impact of EBV status on characteristics and outcomes of posttransplantation lymphoproliferative disorder. Am J Transplant. 2015;15(10):2665-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trappe RU, Dierickx D, Zimmermann H, et al. Response to rituximab induction is a predictive marker in B-cell post-transplant lymphoproliferative disorder and allows successful stratification into rituximab or R-CHOP consolidation in an international, prospective, multicenter phase II trial. J Clin Oncol. 2017;35(5):536-543. [DOI] [PubMed] [Google Scholar]
- 41.Zimmermann H, Koenecke C, Dreyling MH, et al. Modified risk-stratified sequential treatment (subcutaneous rituximab with or without chemotherapy) in B-cell post-transplant lymphoproliferative disorder (PTLD) after solid organ transplantation (SOT): the prospective multicentre phase II PTLD-2 trial. Leukemia. 2022;36(10):2468-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferla V, Rossi FG, Goldaniga MC, Baldini L. Biological difference between Epstein-Barr virus positive and negative post-transplant lymphoproliferative disorders and their clinical impact. Front Oncol. 2020;10:506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gross H, Hennard C, Masouris I, et al. Binding of the heterogeneous ribonucleoprotein K (hnRNP K) to the Epstein-Barr virus nuclear antigen 2 (EBNA2) enhances viral LMP2A expression. PLoS One 2012;7(8):e42106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chapman JR, Bouska AC, Zhang W, et al. EBV-positive HIV-associated diffuse large B cell lymphomas are characterized by JAK/STAT (STAT3) pathway mutations and unique clinicopathologic features. Br J Haematol. 2021;194(5):870-878. [DOI] [PubMed] [Google Scholar]
- 45.Sarkozy C, Hung SS, Chavez EA, et al. Mutational landscape of gray zone lymphoma. Blood. 2021;137(13):1765-1776. [DOI] [PubMed] [Google Scholar]
- 46.Ramis-Zaldivar JE, Gonzalez-Farre B, Nicolae A, et al. MAPK and JAK-STAT pathways dysregulation in plasmablastic lymphoma. Haematologica. 2021;106(10):2682-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Z, Filip I, Gomez K, et al. Genomic characterization of HIV-associated plasmablastic lymphoma identifies pervasive mutations in the JAK-STAT pathway. Blood Cancer Discov. 2020;1(1):112-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leeman-Neill RJ, Soderquist CR, Montanari F, et al. Phenogenomic heterogeneity of post-transplant plasmablastic lymphomas. Haematologica. 2022;107(1):201-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hernández-Verdin I, Kirasic E, Wienand K, et al. Molecular and clinical diversity in primary central nervous system lymphoma. Ann Oncol. 2023;34(2):186-199. [DOI] [PubMed] [Google Scholar]
- 50.Garcia Alvarez HM, Koşaloğlu-Yalçın Z, Peters B, Nielsen M. The role of antigen expression in shaping the repertoire of HLA presented ligands. iScience. 2022;25(9):104975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wells DK, van Buuren MM, Dang KK, et al. Key parameters of tumor epitope immunogenicity revealed through a consortium approach improve neoantigen prediction. Cell. 2020;183(3):818-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Mattos-Arruda L, Vazquez M, Finotello F, et al. Neoantigen prediction and computational perspectives towards clinical benefit: recommendations from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31(8):978-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon reasonable request. The French data privacy authority (CNIL, Commission Nationale de l’Informatique et des Libertés) and European regulations (GDPR, General Data Protection Regulation) do not provide for transmission of the database, nor do the information and consent documents signed by the patients. Consultation by the editorial board or interested researchers of individual participant data that underlie the results reported in the article after deidentification may nevertheless be considered, subject to prior determination of the terms and conditions of such consultation and in respect of compliance with the applicable regulations. For all inquiries, please contact drc-secretariat-promotion@aphp.fr. The R code (version 4.2.1) utilized in our research is accessible upon reasonable request. Please contact the author for correspondence to obtain access.