Abstract
Congenital heart disease (CHD) remains a significant global health concern, affecting approximately 1 % of newborns worldwide. While its accurate causes often remain elusive, a combination of genetic and environmental factors is implicated. In this cross-sectional study, we propose a comprehensive prediction framework leveraging Machine Learning (ML) and Multi-Attribute Decision Making (MADM) techniques to enhance CHD diagnostics and forecasting. Our framework integrates supervised and unsupervised learning methodologies to remove data noise and address imbalanced datasets effectively. Through the utilization of imbalance ensemble methods and clustering algorithms such as K-means, we enhance predictive accuracy, particularly in non-clinical datasets where imbalances are prevalent. Our results demonstrate an improvement of 8 % in recall compared to existing literature, showcasing the efficacy of our approach. Moreover, our framework identifies clusters of patients at the highest risk using MADM techniques, providing insights into susceptibility to CHD. Fuzzy clustering techniques further assess the degree of risk for individuals within each cluster, enabling personalized risk evaluation. Importantly, our analysis reveals that unhealthy lifestyle factors, annual per capita income, nutrition, and folic acid supplementation emerge as crucial predictors of CHD occurrences. Additionally, environmental risk factors and maternal illnesses significantly contribute to the predictive model. These findings underscore the multifactorial nature of CHD development, emphasizing the importance of considering socioeconomic and lifestyle factors alongside medical variables in CHD risk assessment and prevention strategies. Our proposed framework offers a promising avenue for early identification and intervention, potentially mitigating the burden of CHD on affected individuals and healthcare systems globally.
Keywords: Congenital heart disease, Multi-attribute decision-making, Clustering, Risk assessment, Machine learning
1. Introduction
Congenital heart disease (CHD) is the most common type of congenital defect worldwide, affecting approximately 1 % of newborns, making it a prevalent condition that accounts for 28 % of all congenital defects [1,2]. Due to substantial advancements in medical, transcatheter, and surgical treatments for CHD over time, over 95 % of infants born with CHD now survive into adulthood, with over 90 % reaching the age of 40 years or older [3]. The exact causes of CHD in most infants remain unknown. Some infants develop heart defects due to genetic or chromosomal alterations, while others may be influenced by a combination of genetic and environmental factors such as maternal diet, health conditions, or medication use during pregnancy [4,5]. Certain CHDs can be identified before birth through a fetal echocardiogram, a specialized ultrasound that produces images of the developing baby's heart. Early detection of an illness, even before the expected time, can be life-saving [6]. However, some defects may not be detected until after birth or later in life, during childhood or adulthood [7]. CHDs include a wide range of heart abnormalities that differ in seriousness. The particular defect type can greatly impact the disease's development, outlook, and patients' ability to participate in daily tasks [8]. Unrecognized CHDs in infants present a substantial danger of avoidable mortality, sickness, and impairment. Prompt identification of heart issues in infants provides important advantages by allowing for timely assessment and treatment, resulting in quicker evaluations and better clinical conditions [9]. Machine learning (ML) is transforming the healthcare industry by allowing for predictive diagnostics and customized treatment strategies [10]. Supervised learning algorithms use labeled data to make predictions about outcomes, like disease progression or treatment response. Unsupervised learning methods reveal concealed patterns in data without labels, assisting in grouping patients or detecting abnormalities for early detection of diseases. Together, these techniques improve the process of making medical decisions and caring for patients [11,12]. The scientists have investigated the use of ML to forecast prenatal CHD by examining morphological and hemodynamic characteristics from prenatal ultrasound images. Although there has been significant research into this method, building models using ultrasound images presents difficulties [13,14]. Creating a forecasting tool to identify congenital heart disease in babies before birth by analyzing non-clinical information from pregnant women could effectively tackle this problem. Using non-clinical information for forecasting includes lifestyle elements, environmental impacts, and genetic tendencies, enhancing predictive models. Moreover, it enables early identification and custom interventions for improved healthcare results. The primary challenge with non-clinical data in health datasets is imbalanced data distribution, where certain classes or outcomes are significantly underrepresented compared to others. This can lead to biased predictions and hinder the effectiveness of ML models in healthcare applications [15].
The rest of the paper is structured as follows: Section 2 reviews the related works in the field of CHD prediction and management. Section 3 provides a detailed description of the proposed framework. Section 4 presents the results of the research. Section 5 discusses the findings and offers perspectives. Finally, Section 6 provides the concluding remarks and outlines the possible paths for future study.
2. Related works
In recent years, the application of ML and data analysis in healthcare has significantly improved, particularly in the domain of CHD. Numerous studies have explored various approaches to enhance the prediction, diagnosis, and management of CHD, leveraging both clinical and non-clinical data. Kaur and Ahmad [13] proposed a data analysis model to forecast CHD risk and create expectant mother cohorts based on lifestyle similarities. Employing DBSCAN for unsupervised clustering and random forest for prediction, the method achieved 99 % accuracy and outperformed existing approaches, demonstrating the potential of unsupervised learning in refining disease prediction models. Griffeth et al. [16] investigated the impact of preoperative heart failure on outcomes in adult CHD surgeries and developed a 7-feature risk model for postoperative complications using ML, where ejection fraction emerged as the most influential factor. Shi et al. [17] pointed to the development and validation of ML models to predict malnutrition in children with CHD post-surgery, utilizing explainable ML methods for insight. The study enrolled 536 children, with XGBoost showing superior predictive performance, highlighting crucial features like postoperative weight scores for early intervention strategies. Junior et al. [18] suggested an ML model to forecast ICU stays post-surgery for CHD patients, aiming to improve care planning. Analyzing data from 2240 patients in a Brazilian hospital, the Light Gradient Boosting Machine achieved a low error rate, while the CatBoost Classifier showed promising accuracy. Xu et al. [19] proposed an ML system for classifying 17 categories of CHD types, achieving human-expert-level performance. This system, trained on a large dataset from various CT machines, offers high accuracy (86.03 %) and sensitivity (82.91 %), enhancing CHD diagnosis globally, particularly in regions with limited expert radiologists. Luo et al. [20] recommended examining the characteristics of pre-closure and their impact on post-closure outcomes in patients with uncorrected isolated simple shunts and pulmonary arterial hypertension. Using unsupervised and supervised ML, they analyzed patient data, identifying key features and clustering patterns. While certain characteristics correlated with better post-closure outcomes, supervised learning models did not effectively predict these outcomes. He et al. [21] proposed a predictive model for prolonged mechanical ventilation (PMV) in congenital heart disease (CHD) patients with airway stenosis (AS) managed conservatively. The model identified key risk factors, including low weight, complex CHD, and tracheobronchomalacia, with strong predictive accuracy. Early identification and routine surveillance of high-risk patients are essential for improving the outcomes. Reddy et al. [22] suggested that there have been significant advancements in prenatal diagnosis and management of CHD, with fetal echocardiography achieving an 85 % accuracy rate in detecting cardiac anomalies. Although prenatal CHD diagnosis improves risk assessment and patient outcomes, there is a need to enhance identification rates through increased training and the integration of artificial intelligence (AI) to optimize image acquisition, measurements, diagnosis, and outcome prediction, despite existing barriers to widespread adoption. The summary of key studies in the literature, highlighting the objectives, methodologies, and findings, is presented in Table 1.
Table 1.
Summary of key studies on CHD prediction and management.
| Study | Purpose | Methodology | Dataset Type | Individual Risk Assessment | Outcomes |
|---|---|---|---|---|---|
| Kaur and Ahmad [13] | Predict CHD risk in expectant mothers and create cohorts based on lifestyle using population-based cross-sectional data | DBSCAN for clustering, Random Forest for prediction; k-NN for imputation; SMOTE for balancing data | non-clinical data | No | Achieved 99 % accuracy and 0.91 AUC. Clustering revealed complex factors affecting prediction. |
| Griffeth et al. [16] | Evaluate the impact of preoperative heart failure on reoperative cardiac surgery outcomes in adult CHD patients | Retrospective cohort study, Multivariable logistic regression, Gradient Boosting, SHAP for feature importance | Clinical data | No | Developed a 7-feature risk model with an AUC of 0.76; identified ejection fraction as the most influential factor. |
| Shi et al. [17] | Predict malnutrition in children with CHD post-surgery | Prospective cohort study, XGBoost, and other ML models, SHAP for feature importance | Clinical data | No | XGBoost showed superior predictive performance with early intervention potential. Key features identified using SHAP. |
| Junior et al. [18] | Develop a predictive model for ICU length of stay (LOS) after CHD surgery | Light Gradient Boosting Machine for regression, CatBoost Classifier for clustering | Clinical data | No | LightGBM achieved a low Mean Squared Error, while the CatBoost Classifier showed high accuracy and AUC. Key predictors included mechanical ventilation duration and weight. |
| Xu et al. [19] | Classify 17 categories of CHD types using AI | AI system combining deep learning and ML for feature extraction and classification | Clinical CT images | No | Achieved 86.03 % accuracy comparable to junior radiologists; high sensitivity (82.91 %) and potential for clinical integration. |
| Luo et al. [20] | Investigate preclosure characteristics and postclosure outcomes in CHD patients with simple shunts associated with PAH | Unsupervised and supervised ML for clustering and model construction | Clinical data | No | Identified clusters with significantly different postclosure outcomes; supervised models underperformed. |
| He et al. [21] | Predict prolonged mechanical ventilation in CHD patients with airway stenosis | Retrospective study, Predictive modeling using ROC curve analysis | Clinical data | No | AUC of 0.847; identified weight, CPB duration, and complex CHD as key risk factors. Divided patients into high and low-risk groups. |
| Reddy et al. [22] | Enhance prenatal diagnosis and management of CHD using fetal echocardiography | Review and proposal for AI integration in fetal echocardiography | Clinical data | No | Fetal echocardiography achieved 85 % accuracy; AI proposed to improve detection and diagnosis accuracy, with potential integration into clinical practice. |
While the existing literature offers several approaches for predicting the cases of CHD, there remain notable gaps within these methodologies.
-
•
Most studies concentrate on using clinical and accurate data to predict cases of CHD, overlooking the potential of non-clinical data in forecasting CHD incidences prior to childbirth. This discrepancy highlights the necessity for effective models that can integrate various data sources to improve predictive accuracy and detect CHD cases early.
-
•
Another significant gap lies in the lack of comprehensive categorization of patients based on their susceptibility to CHD, utilizing key features extracted from CHD datasets. By establishing distinct categories and defining the significance of each category in relation to CHD risk, healthcare practitioners can better tailor preventive measures and interventions for at-risk individuals.
-
•
Additionally, there is a significant gap in the available studies regarding the measurement of CHD risk and the development of a plan to reduce these risks.
Addressing these gaps holds the potential to greatly enhance our understanding of early CHD prediction and risk mitigation for individuals even before birth. Consequently, this paper's key contributions can be summarized as follows.
-
i.
A comprehensive prediction framework is presented, designed to not only enhance accuracy but also effectively determine cases of CHD in forecasting. This framework demonstrates proficiency in navigating noise and addressing imbalanced data by integrating supervised and unsupervised learning methodologies.
-
ii.
By leveraging feature importance metrics and employing Multi-Attribute Decision-Making (MADM) techniques, the framework identifies clusters of patients at the highest risk, prioritizing those most in danger within the population.
-
iii.
Utilizing fuzzy clustering techniques, the framework assesses the degree of risk within each identified cluster, offering insights into susceptibility to CHD. Moreover, it identifies crucial features instrumental in mitigating these risks, thus facilitating targeted interventions for risk reduction.
3. Requirements and framework
In this section, we introduce the requirements methods, laying the groundwork for the proposed framework. We begin by providing a comprehensive description of the dataset utilized in this study, offering insights into its structure and key characteristics. Following this, we provide an overview of the approaches employed, setting the stage for a detailed discussion on preprocessing methods. Finally, we delve into an in-depth examination of the framework, ending with a comprehensive understanding of its design and implementation. All data preprocessing, clustering, and classifier evaluations were conducted using Python.
3.1. Dataset description
This study utilized data from a large-scale, retrospective epidemiological survey conducted by the Population and Family Planning Commission (PFPC) of Shanxi Province, China, between 2006 and 2008. The survey covered six counties—Pingding, Dai, Fenyang, Huairen, Zhongyang, and Jiaokou—selected through stratified random cluster sampling. The data were collected for 36,300 live infants and their mothers, with 78 cases of CHD identified, making the dataset highly unbalanced. The ethical approval was obtained from the Human Research Ethics Committee of Shanxi PFPC. The data included comprehensive maternal demographic and health information gathered via questionnaires. For this study, nine key risk factors are selected based on prior research, resulting in a final dataset of 10 variables including 9 predictors and 1 binary outcome variable (CHD presence) [23]. Table 2 provides a concise overview of these features. For a comprehensive understanding of the dataset and to grasp the significance of each feature item, readers are encouraged to refer to the detailed description provided by Luo et al. [24].
Table 2.
Summary of dataset features.
| Feature | Description | Range |
|---|---|---|
| Maternal delivery age | Maternal delivery age ≥30 | 0–1 |
| Annual per capita income | less than 1000(¥) | 1–5 |
| 1000–2000¥ | ||
| 2000–4000¥ | ||
| 4000–8000¥ more than 8000¥ | ||
| Family history | Parental consanguinity | 0–2 |
| Birth defects in immediate family members | ||
| Birth defects in previous infants | ||
| Maternal previous illness history | Hepatitis | 0–6 |
| Epilepsy | ||
| Anemia | ||
| Diabetes | ||
| Heart disease Spontaneous abortion | ||
| Thyroid disease | ||
| Other | ||
| Nutrition and folic acid supplementation | Vegetable deficiency | 0–5 |
| Meat deficiency | ||
| Folic acid deficiency | ||
| Maternal illness | Cold | 0–6 |
| Fever | ||
| Threatened abortion | ||
| Reproductive tract infections | ||
| Hyperemesis gravidarum | ||
| Rash and fever | ||
| Other | ||
| Medication use | Cold medicines | 0–7 |
| Antiemetic | ||
| Antibiotic | ||
| Antiepileptic | ||
| Sedative | ||
| Contraceptive | ||
| Abortion prevention agent | ||
| Other | ||
| Environmental exposures of risk factors | Pesticides | 0–6 |
| Chemical fertilizers | ||
| X-rays | ||
| Computer use | ||
| Pets | ||
| Pollution source in the area of residence | ||
| Unhealthy lifestyle | Periconceptional smoking | 0–8 |
| Family member smoking | ||
| Periconceptional drinking | ||
| Family member drinking |
3.2. Clustering
Clustering serves as a powerful technique for grouping similar data points based on specific features, aiming to uncover the underlying patterns or structures within the dataset. In this paper, two distinct clustering methods are employed. The first method, K-means, is utilized within a predictive model, where the optimal number of clusters is determined using the silhouette and Davies-Bouldin Index (DBI) methods. K-means is selected for its simplicity, speed, efficiency, and widespread adoption, facilitating the categorization of data into cohesive groups. While other clustering methods could be employed, K-means is preferred due to its practical advantages. The second clustering method utilized is fuzzy c-means clustering, employed for individual risk evaluation. This method assigns probabilities to each instance, indicating their likelihood of belonging to each cluster. These probabilities serve as crucial tools in defining the risk of CHD, offering insights into individual risk factors, and facilitating more personalized risk assessment.
3.2.1. K-means clustering
The K-means algorithm, developed by MacQueen in 1967 [25], is a popular clustering method that provides a systematic way to classify data points according to their similarity. The process starts by selecting a fixed number of clusters, denoted as K. Then, K cluster centroids are randomly placed throughout the dataset. After that, every data point is allocated to the closest centroid based on distance measurements. Centroids are recalculated throughout the assignment by averaging the positions of the data points in each cluster. This repetitive procedure goes on until convergence, with the objective of reducing the total squared distances between data points and their corresponding centroids. This technique, known for its straightforwardness and success, continues to be a foundational element in many clustering scenarios [26].
3.2.2. Fuzzy C-means (FCM)
Fuzzy clustering represents an efficient approach in unsupervised data analysis and model development, offering a departure from rigid class assignments by allowing objects to exhibit partial memberships ranging from 0 to 1 rather than enforcing strict categorization. Among the most prominent methods in this domain is the FCM algorithm, pioneered by Bezdek et al., in 1984 [27]. FCM assigns each data point a membership grade for each cluster center, indicating its degree of association on a continuum between 0 and 1, based on proximity measures. The closer a data point lies to a cluster center, the higher its membership value for that cluster, with the sum of memberships for each point equating to one. Following each iteration, memberships and cluster centers undergo updates according to a defined formula [28]. This iterative process enables FCM to effectively delineate complex data patterns while accommodating varying degrees of data point association with different clusters.
3.2.3. DBI technique
DBI was created by Davies and Bouldin in 1979 [29] as a means to assess the effectiveness of clustering. The evaluation of cluster effectiveness involves examining the similarity within each cluster as well as the spread across clusters. DBI stands out from other metrics because lower values signify clearer distinctions between clusters, making it easier to interpret. DBI takes into consideration both the density of clusters and the distance between clusters without assuming anything about their shapes or densities [30]. Therefore, DBI is identified as a flexible and trustworthy tool for evaluating clustering performance on different datasets.
3.2.4. Silhouette technique
The silhouette method [31] is a useful tool for finding the best number of clusters when using the K-means algorithm. Initially, it computes the average distance between a data point and all others in the same cluster, known as the intra-cluster distance. Afterward, the inter-cluster distance is calculated as the average distance between the data point and the other points in the nearest neighboring cluster. The silhouette coefficient is obtained by calculating the ratio of the difference between these two distances, divided by the higher of the two distances. This scale, which goes from −1 to 1, indicates how closely a data point matches others in its cluster. A score of 1 indicates significant similarity within the cluster and clear dissimilarity from other clusters, whereas a score of −1 represents the opposite, indicating low similarity within its designated cluster and significant similarity to points in other clusters [32].
3.3. MADM techniques
In this study, the MADM technique known as Combined Compromise Solution (CoCoSo) is employed to discern the distinctions between clusters. CoCoSo integrates a simple additive weighting and exponentially weighted product model, offering a comprehensive set of compromise solutions. This technique is utilized to facilitate decision-making by considering multiple attributes simultaneously. The weights assigned in CoCoSo are derived from the importance of the features of the predictive model. This approach prioritizes important features, assigning them higher weights in ranking the clusters, thereby enhancing the differentiation between them. A detailed explanation of the CoCoSo technique can be found in reference [33].
3.4. Preprocessing
Preprocessing is crucial for ensuring the data quality and reliability, as it addresses the missing values and standardizes the features for accurate analysis. Table 3 provides an overview of the number of missing values in each feature, guiding the necessary preprocessing steps.
Table 3.
Number of missing values across features in the CHD dataset.
| Feature | Missing value |
|---|---|
| Annual per capita income | 796 |
| Congenital Heart Defects | 0 |
| Maternal delivery age | 241 |
| Family history | 996 |
| Maternal previous illness history | 229 |
| Nutrition and folic acid supplementation acid supplementation | 172 |
| Maternal illness | 241 |
| Medication use | 201 |
| Environmental exposures of risk factors factors | 29 |
| Unhealthy lifestyle | 56 |
In the preprocessing phase, the first step involves filtering out instances with more than 50 % missing values, as these may skew the analysis. The remaining missing values are then imputed using the mode of their respective columns. Given the integer nature of the data, where each value represents the number of factors associated with each instance, utilizing the mode for imputation is logical and maintains the integrity of the dataset. While the mode imputation can disproportionately represent the majority class, potentially skewing the data and affecting the model's performance, the low percentage of missing values in this dataset—ranging between 0 % and 2.7%—mitigates this concern, making mode imputation an acceptable approach in this context. Additionally, standardization is performed before employing the data in prediction and clustering tasks. This ensures that all variables are on a comparable scale, preventing any single feature from dominating the analysis due to differences in magnitude.
3.5. Proposed framework
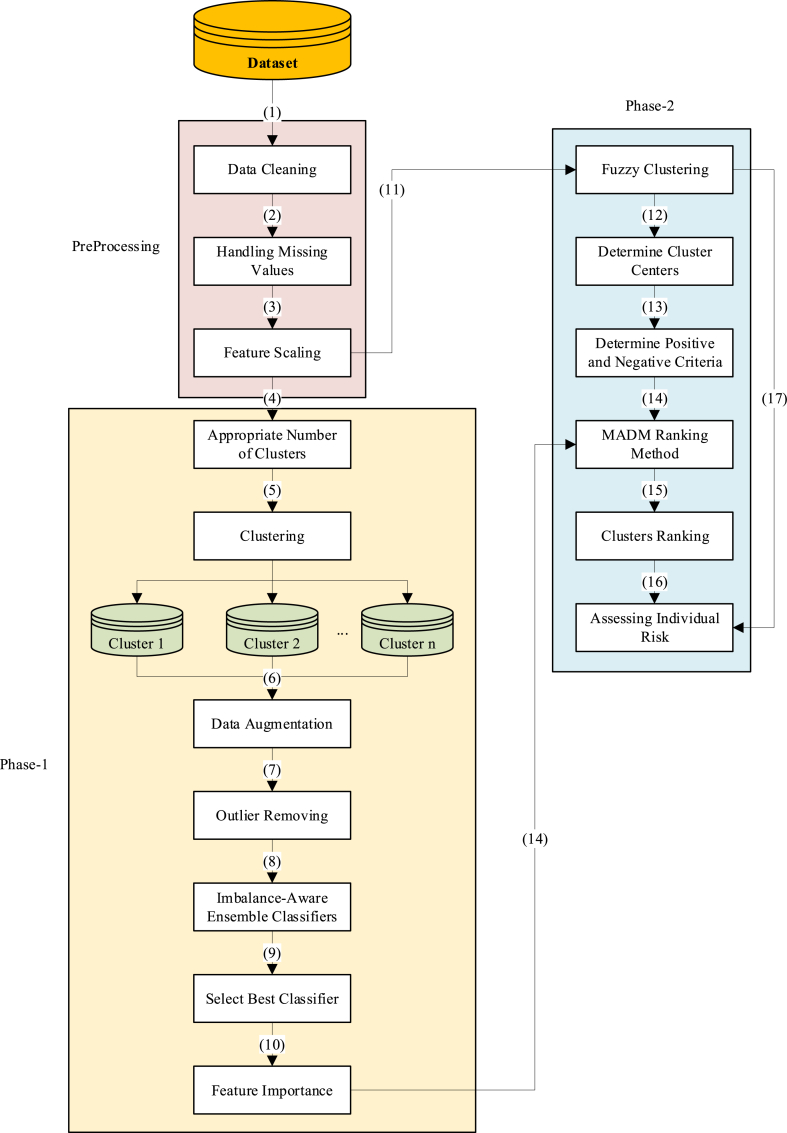
The proposed framework is structured into two phases, following preprocessing. Phase 1 is dedicated to accurately predicting cases of CHD, while phase 2 emphasizes assessing individual CHD risk levels. The framework commences with preprocessing steps 1 to 3, followed by acquiring an appropriate cluster number using silhouette and DBI methods in step 4. Subsequently, K-means clustering is employed to segment the data into multiple clusters. The motivation behind utilizing clustering techniques in this classification task stems from the dataset's high degree of imbalance and the necessity for more effective oversampling. While the data is labeled, clustering helps to group similar instances together, thereby allowing the generation of more meaningful and contextually relevant synthetic instances during the oversampling process. By working within clusters, we ensure that the newly generated instances are more representative of the underlying data distribution, which improves the classifier's ability to differentiate between classes. In step 6, the clusters are evaluated to ensure an adequate representation of the minority class. If any cluster lacks sufficient minority class instances, their data are randomly duplicated to augment their numbers. Following this, step 7 involves removing outliers from the majority class by eliminating a percentage of the farthest instances from each cluster center. Given the data's pronounced imbalance, in step 8, traditional classifiers may not yield optimal results. Hence, ensemble models integrated with imbalance correction techniques such as EasyEnsembleClassifier [34], BalancedRandomForestClassifier [35], BalancedBaggingClassifier [36], and RUSBoostClassifier [37] are employed. Step 9 involves identifying the best classifier, while step 10 focuses on deriving feature importance from this classifier to further enhance the model's performance.
In the second phase, utilizing the preprocessed data from step 11, fuzzy clustering is conducted using the c-means method. Subsequently, in step 12, the centers of these clusters are utilized as alternatives for MADM ranking. The objective is to rank the clusters based on the risk they pose for individuals. In step 13, positive and negative criteria are determined, followed by MADM ranking using the CoCoSo method in step 14. This process incorporates both the criteria and the feature importance derived from the classifier as weights. After ranking the clusters and calculating scores for each cluster in step 16, the probability of each instance belonging to each cluster from fuzzy clustering is determined in step 17. This information enables the computation of Individual Risk, as outlined in Equation (1). Fig. 1 illustrates the structure and steps of the proposed framework.
| (1) |
where represents the probability of assigning each instance to cluster , denotes the score of cluster , and signifies the number of clusters.
Fig. 1.
The proposed framework.
4. Results
In this section, we delve into the outcomes of the proposed framework. Initially, we explore the results pertaining to phase 1, focusing on its predictive capabilities regarding CHD. Subsequently, we scrutinize the phase-2 results to assess the framework's efficacy in evaluating individual CHD risk levels.
4.1. Phase-1
After obtaining the preprocessed dataset suitable for analysis, we employ two optimal cluster-finding methods, namely silhouette and DBI, to determine the appropriate number of clusters. These methods investigate the clusters within the range of 3–15. The average silhouette and DBI results suggest that 7 clusters are appropriate for the dataset. Clinically, the identification of 7 distinct clusters holds significant relevance as it allows for the stratification of patients based on varying levels of CHD risk. Each cluster represents a group with unique characteristics and risk profiles. After clustering, it is important to ensure that each cluster has enough minority instances (CHD cases) for the effective use of imbalance ensemble methods. To achieve this, the clusters with fewer than 20 minority instances are duplicated randomly until they reach at least 20 instances. The threshold of 20 is chosen based on empirical testing to ensure that the ensemble models perform optimally; it is not aimed at balancing the clusters, as the number of majority instances in each cluster ranges from 227 to 12,966. This duplication step is considered to ensure that the ensemble models operate effectively within each cluster, rather than to achieve balance between majority and minority classes. Identifying and eliminating outliers from the dataset can help the model become more robust and enhance its performance. In this scenario, outliers are eliminated from the majority class (non-CHD cases) because of the dataset's high-class imbalance. The distance of each instance from the center of the cluster is computed, and a percentage of the farthest instances is subsequently eliminated for every cluster. In this dataset, a 20 % elimination rate is employed. Table 4 presents the performance of four classifiers evaluated using four metrics: accuracy, F1 score, recall, and precision.
Table 4.
Evaluation of ensemble classifiers’ performance across accuracy, F1 score, recall, and precision.
| Classifier | Accuracy | F1 Score | Recall | Precision |
|---|---|---|---|---|
| BalancedBaggingClassifier | 0.857 | 0.227 | 0.839 | 0.167 |
| BalancedRandomForestClassifier | 0.830 | 0.228 | 0.886 | 0.159 |
| EasyEnsembleClassifier | 0.794 | 0.168 | 0.897 | 0.116 |
| RUSBoostClassifier | 0.830 | 0.126 | 0.592 | 0.093 |
This table displays the performance metrics of four classifiers: BalancedBaggingClassifier, BalancedRandomForestClassifier, EasyEnsembleClassifier, and RUSBoostClassifier. Notably, in the healthcare dataset with a high degree of class imbalance, it is crucial to enhance the Recall score to achieve satisfactory results on positive cases. In comparison to a previous study [13] which attained a recall of 0.8, our models demonstrate varying degrees of success. Particularly, the EasyEnsembleClassifier and BalancedRandomForestClassifier exhibit notable recall scores of 0.897 and 0.886, respectively. For a detailed breakdown of results on each cluster, please refer to Appendix A.
Utilizing the BalancedRandomForestClassifier as the best-performing classifier, the importance of each feature is derived from the model. These feature importances serve as weights for the MADM ranking method. Although the EasyEnsembleClassifier demonstrates slightly better recall performance compared to the BalancedRandomForestClassifier, the superior performance of the BalancedRandomForestClassifier across other metrics justifies its selection as the best-performing classifier.
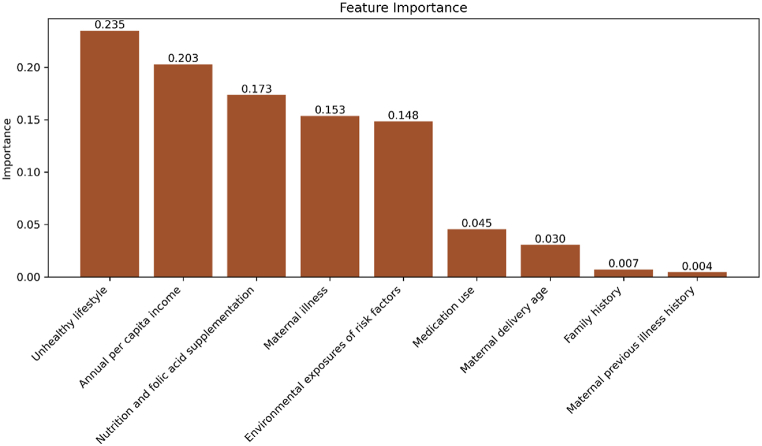
Fig. 2 displays the importance of features that predict cases of CHD. Unhealthy lifestyle and annual per capita income are the most important predictors of CHD occurrences, with nutrition and folic acid supplementation also playing significant roles. Environmental risk factors and maternal illnesses are also significantly important in the predictive model. These results highlight the multiple factors involved in the development of CHD, stressing the need to consider socioeconomic and lifestyle factors, in addition to medical issues, when assessing and preventing CHD risks.
Fig. 2.
Feature importance of the BalancedRandomForestClassifier.
4.2. Phase-2
In phase 2, the fuzzy clustering method, specifically c-means clustering, is employed. This clustering method allows us to obtain the probability of each individual belonging to each cluster. Following clustering, the cluster centers are utilized as alternatives and are presented in Table 6.
Table 6.
Cluster centers derived from C-Mean clustering.
| Cluster | Annual per capita income | Environmental exposures of risk factors | Family history | Maternal delivery age | Maternal illness | Maternal previous illness history | Medication use | Nutrition and folic acid supplementation | Unhealthy lifestyle |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 3.533 | 0.206 | 0.006 | 0.000 | 0.119 | 0.028 | 0.032 | 2.346 | 1.504 |
| 1 | 2.918 | 0.152 | 0.009 | 0.000 | 0.133 | 0.029 | 0.027 | 3.956 | 0.244 |
| 2 | 2.600 | 2.616 | 0.010 | 0.184 | 0.256 | 0.059 | 0.154 | 3.087 | 2.023 |
| 3 | 1.647 | 0.279 | 0.009 | 0.000 | 0.156 | 0.034 | 0.036 | 2.695 | 1.757 |
| 4 | 2.863 | 0.267 | 0.008 | 1.000 | 0.155 | 0.065 | 0.043 | 3.046 | 1.507 |
| 5 | 3.458 | 0.330 | 0.005 | 0.023 | 0.265 | 0.047 | 0.043 | 4.146 | 2.547 |
| 6 | 2.722 | 0.873 | 0.012 | 0.166 | 1.370 | 0.131 | 1.579 | 3.182 | 1.797 |
To categorize the criteria into positive and negative, we defined positive criteria as those where a higher value correlates with a worse outcome, thereby increasing the risk of CHD. Conversely, negative criteria are those where higher values may mitigate CHD risk. In this context, all criteria except “Annual per capita income” are considered positive, as their higher values are associated with increased CHD risk. The “Annual per capita income” is deemed a negative criterion because higher income levels are generally associated with better access to healthcare, improved living conditions, and healthier lifestyles, all of which can contribute to reducing the risk of CHD. This categorization is based on existing literature and socioeconomic research, which consistently shows that individuals with higher income levels tend to have lower health risks, including CHD. Thus, this criterion is defined negatively in this paper, as higher income is expected to mitigate the risk rather than exacerbate it. The impact of this categorization on the results is significant, as it influences the weights and rankings derived from the CoCoSo method, which incorporates the feature importances in evaluating each cluster. Table 5 presents the scores of each cluster.
Table 5.
The cluster scores obtained from the CoCoSo MADM method.
| Cluster | Score |
|---|---|
| 0 | 1.314866 |
| 1 | 1.552812 |
| 2 | 3.161115 |
| 3 | 2.564732 |
| 4 | 2.576163 |
| 5 | 2.772779 |
| 6 | 3.237092 |
The MADM scores presented in Table 5 represent the risk of CHD for each cluster. A higher score indicates a greater risk of CHD within the respective cluster. Cluster 2 and Cluster 6 exhibit the highest scores, suggesting they are associated with elevated CHD risk levels. Conversely, Cluster 0 has the lowest score, indicating a relatively lower risk of CHD within this cluster. These scores offer valuable insights into the varying degrees of CHD risk across different clusters, aiding in targeted interventions and risk management strategies.
For each individual, the evaluation of individual risk involves leveraging both the cluster scores and the probability of belonging to each cluster. By considering these factors, the individual risk of CHD can be efficiently computed. A higher individual risk signifies an elevated likelihood of CHD and indicates the need for intensified interventions and a reassessment of the individual's situation. This holistic approach incorporates both the individual's cluster memberships and the associated risk levels, providing a comprehensive assessment of CHD risk.
5. Discussion
CHD is the predominant congenital defect globally, with the specific origins in most babies still unidentified. Quickly identifying heart problems in infants is crucial for timely treatment, leading to faster evaluations and improved clinical outcomes. This study introduces a two-phase approach to predict non-clinical congenital heart disease data efficiently. The first phase utilizes a combination of clustering and ensemble classifiers to predict imbalanced CHD data. In the second phase, fuzzy clustering and MADM are used together to enhance the framework for assessing individual risk. A key aspect of this study is the emphasis on recall, which measures the ability of a model to correctly identify positive cases. In the context of CHD and other critical diseases, recall is arguably more important than accuracy because it ensures that high-risk cases are not overlooked. An existing study mentioned in the literature survey reported a 99 % accuracy but only 80 % recall. Although our approach does not achieve the same level of accuracy, it does attain a higher recall of approximately 90 %. This improvement in recall indicates that our model is better at identifying true positive cases, which is crucial in medical diagnostics where missing a positive case can have serious consequences. The discrepancy between high accuracy and lower recall in the existing study could be due to a model's potential bias towards the majority class, which is often the case in imbalanced datasets. High accuracy might indicate that the model correctly identifies the majority of non-risk cases but fails to adequately detect high-risk cases, which is where recall becomes critical.
In the second phase of the study, the focus shifts to calculating the individual risk, which is derived from two key components; the fuzzy cluster memberships and the associated cluster scores. The significance of the MADM technique, specifically the CoCoSo technique, lies in its ability to calculate a comprehensive risk score for each cluster based on its features. This approach ensures that the risk assessment for each cluster is calculated according to the most critical factors influencing CHD. In addition, by integrating the fuzzy probability of an individual belonging to each cluster, the MADM technique facilitates the calculation of a personalized, weighted risk score for each person. This dual-layered approach enhances the precision of individual risk assessments, enabling more targeted and effective intervention strategies.
This framework highlights the significance of non-clinical data and the impact of socioeconomic and lifestyle elements. Focusing more on this area and incorporating additional features and details into the data could lead to more precise predictions and prevent CHD from occurring. With the individual risk outlined in this document, one can assess the level of danger for each individual and advise them on how to improve these factors to reduce the risk of CHD.
6. Conclusions
CHD is a major public health issue, impacting many infants worldwide and creating significant challenges for healthcare systems and affected families. Despite significant progress in medical research, the causes of CHD are still numerous and complex, requiring comprehensive strategies for forecasting and avoiding it. In this research, we introduced a comprehensive predictive framework that utilizes ML to detect CHD cases in unborn babies through the analysis of non-clinical information from pregnant women. Our approach combines supervised and unsupervised learning methods by incorporating MADM and fuzzy clustering techniques to handle imbalanced datasets and noisy non-clinical data effectively. By conducting thorough experiments, we have proven that ensemble methods, especially when paired with clustering algorithms, are successful in enhancing predictive precision and detecting subtle patterns in the data. Significantly, the combination of K-means clustering with BalancedRandomForestClassifier exhibited superior performance compared to other ensembles, yielding an 8 % improvement in recall over existing literature. Additionally, Examination of the importance of features has given important perspectives on the factors influencing the occurrence of CHD, where socioeconomic determinants, maternal health indicators, and environmental exposures have been identified as significant predictors. Understanding the complex relationship between lifestyle factors like diet and financial status, as well as genetic tendencies, highlights the intricate nature of CHD development and emphasizes the significance of comprehensive risk assessment methods.
Nevertheless, our research has some limitations and numerous opportunities for more investigation and improvement. Relying on non-clinical data presents inherent difficulties since these datasets might not encompass all the variables that influence CHD risk. Incorporating clinical data, which may contain in-depth medical backgrounds and diagnostic details, can improve the predictive abilities of our model and offer a more complete insight into the causes of CHD. The future research should focus on collecting data from a diverse range of demographic groups to ensure that predictive models can be effectively used across various contexts. This is important because cultural differences, dietary habits, genetic factors, and socio-economic conditions can all influence the CHD risk in different populations. By including a broader spectrum of non-clinical data, one can develop more accurate and universally applicable predictive models that account for the unique characteristics of each population. Comparing different geographic areas could help explain differences in CHD risk factors and guide the development of interventions tailored to each region which is suggested for future research. Moreover, improvements in data augmentation methods and ensemble learning strategies provide hopeful solutions for tackling data imbalance and improving the predictive power of models.
CRediT authorship contribution statement
Amirreza Salehi: Writing – original draft, Visualization, Validation, Project administration, Methodology, Investigation, Data curation, Conceptualization. Majid Khedmati: Writing – review & editing, Validation, Supervision, Project administration, Conceptualization.
Ethical and informed consent for data used
Ethical approval was not required for this study as the data used are publicly available. Informed consent was obtained by the original data collectors or is not applicable as the data are anonymized and publicly accessible.
Data availability and access
Sharing research data helps other researchers evaluate your findings, build on your work and to increase trust in your article. We encourage all our authors to make as much of their data publicly available as reasonably possible. Please note that your response to the following questions regarding the public data availability and the reasons for potentially not making data available will be available alongside your article upon publication.
Has data associated with your study been deposited into a publicly available repository?
-
•
Yes
Sharing research data helps other researchers evaluate your findings, build on your work and to increase trust in your article. We encourage all our authors to make as much of their data publicly available as reasonably possible. Please note that your response to the following questions regarding the public data availability and the reasons for potentially not making data available will be available alongside your article upon publication.
Has data associated with your study been deposited into a publicly available repository?
-
•
The data referenced in this manuscript are publicly available and can be accessed through [24]. The dataset utilized in this study comprises non-clinical data spanning six counties in China from 2006 to 2008. It consists of 36,300 instances and encompasses 10 distinct features. Also, we provide the dataset in the GitHub: https://github.com/Amir27Salehi/Congenital-Heart-Disease-
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Amirreza Salehi, Email: Sar.salehiamiri@ie.sharif.edu.
Majid Khedmati, Email: khedmati@sharif.edu.
Appendix A.
The comprehensive results of the four classifiers on each cluster are elaborated in Table A1.
Table A1.
Evaluation of ensemble classifiers' performance across clusters
| Cluster | Classifier | Accuracy | F1 Score | Recall | Precision |
|---|---|---|---|---|---|
| 1 | EasyEnsembleClassifier | 0.815 | 0.009 | 1.000 | 0.005 |
| 1 | RUSBoostClassifier | 0.869 | 0.013 | 1.000 | 0.007 |
| 1 | BalancedBaggingClassifier | 0.799 | 0.009 | 1.000 | 0.004 |
| 1 | BalancedRandomForestClassifier | 0.861 | 0.013 | 1.000 | 0.006 |
| 4 | EasyEnsembleClassifier | 0.926 | 0.154 | 1.000 | 0.083 |
| 4 | RUSBoostClassifier | 0.937 | 0.176 | 1.000 | 0.097 |
| 4 | BalancedBaggingClassifier | 0.955 | 0.231 | 1.000 | 0.130 |
| 4 | BalancedRandomForestClassifier | 0.944 | 0.194 | 1.000 | 0.107 |
| 0 | EasyEnsembleClassifier | 0.452 | 0.003 | 0.667 | 0.002 |
| 0 | RUSBoostClassifier | 0.605 | 0.000 | 0.000 | 0.000 |
| 0 | BalancedBaggingClassifier | 0.827 | 0.011 | 0.667 | 0.006 |
| 0 | BalancedRandomForestClassifier | 0.750 | 0.008 | 0.667 | 0.004 |
| 6 | EasyEnsembleClassifier | 0.978 | 0.308 | 1.000 | 0.182 |
| 6 | RUSBoostClassifier | 0.973 | 0.267 | 1.000 | 0.154 |
| 6 | BalancedBaggingClassifier | 0.995 | 0.667 | 1.000 | 0.500 |
| 6 | BalancedRandomForestClassifier | 0.995 | 0.667 | 1.000 | 0.500 |
| 5 | EasyEnsembleClassifier | 0.872 | 0.150 | 1.000 | 0.081 |
| 5 | RUSBoostClassifier | 0.917 | 0.154 | 0.667 | 0.087 |
| 5 | BalancedBaggingClassifier | 0.929 | 0.174 | 0.667 | 0.100 |
| 5 | BalancedRandomForestClassifier | 0.947 | 0.300 | 1.000 | 0.176 |
| 3 | EasyEnsembleClassifier | 0.839 | 0.033 | 1.000 | 0.017 |
| 3 | RUSBoostClassifier | 0.875 | 0.011 | 0.250 | 0.006 |
| 3 | BalancedBaggingClassifier | 0.840 | 0.033 | 1.000 | 0.017 |
| 3 | BalancedRandomForestClassifier | 0.789 | 0.025 | 1.000 | 0.013 |
| 2 | EasyEnsembleClassifier | 0.674 | 0.516 | 0.615 | 0.444 |
| 2 | RUSBoostClassifier | 0.630 | 0.261 | 0.231 | 0.300 |
| 2 | BalancedBaggingClassifier | 0.652 | 0.467 | 0.538 | 0.412 |
| 2 | BalancedRandomForestClassifier | 0.522 | 0.389 | 0.538 | 0.304 |
References
- 1.Han P.-L., Jiang L., Cheng J.-L., Shi K., Huang S., Jiang Y., Jiang L., Xia Q., Li Y.-Y., Zhu M. Artificial intelligence-assisted diagnosis of congenital heart disease and associated pulmonary arterial hypertension from chest radiographs: a multi-reader multi-case study. Eur. J. Radiol. 2024;171 doi: 10.1016/j.ejrad.2023.111277. [DOI] [PubMed] [Google Scholar]
- 2.Boneva R.S., Botto L.D., Moore C.A., Yang Q., Correa A., Erickson J.D. Mortality associated with congenital heart defects in the United States: trends and racial disparities, 1979–1997. Circulation. 2001;103(19):2376–2381. doi: 10.1161/01.cir.103.19.2376. [DOI] [PubMed] [Google Scholar]
- 3.Egbe A.C., Miranda W.R., Ahmed M., Karnakoti S., Kandlakunta S., Eltony M., Meshreky M., Burchill L.J., Connolly H.M. Incidence and correlates of mortality in adults with congenital heart disease of different age groups. International Journal of Cardiology Congenital Heart Disease. 2024 [Google Scholar]
- 4.Patel S.S., Burns T.L. Nongenetic risk factors and congenital heart defects. Pediatr. Cardiol. 2013;34:1535–1555. doi: 10.1007/s00246-013-0775-4. [DOI] [PubMed] [Google Scholar]
- 5.Jenkins K.J., Correa A., Feinstein J.A., Botto L., Britt A.E., Daniels S.R., Elixson M., Warnes C.A., Webb C.L. Noninherited risk factors and congenital cardiovascular defects: current knowledge: a scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115(23):2995–3014. doi: 10.1161/CIRCULATIONAHA.106.183216. [DOI] [PubMed] [Google Scholar]
- 6.Diwakar M., Tripathi A., Joshi K., Memoria M., Singh P. Latest trends on heart disease prediction using machine learning and image fusion. Mater. Today: Proc. 2021;37:3213–3218. [Google Scholar]
- 7.Prevention, C. f. D. C. a. 2023. What are CHDs.https://www.cdc.gov/ncbddd/heartdefects/facts.html#:∼:text=CHDs%20are%20present%20at%20birth,formed%20parts%20of%20the%20heart [Google Scholar]
- 8.Moons P., Van Deyk K., De Geest S., Gewillig M., Budts W. Is the severity of congenital heart disease associated with the quality of life and perceived health of adult patients? Heart. 2005;91(9):1193–1198. doi: 10.1136/hrt.2004.042234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richmond S., Wren C. Early diagnosis of congenital heart disease. Semin. Neonatol. 2001;6(1):27–35. doi: 10.1053/siny.2000.0028. [DOI] [PubMed] [Google Scholar]
- 10.Kaur I., Ahmad T., Doja M. A systematic review of medical expert systems for cardiac arrest prediction. Curr. Bioinf. 2024;19(6):551–570. [Google Scholar]
- 11.Miller R.J., Bednarski B.P., Pieszko K., Kwiecinski J., Williams M.C., Shanbhag A., Liang J.X., Huang C., Sharir T., Hauser M.T. Clinical phenotypes among patients with normal cardiac perfusion using unsupervised learning: a retrospective observational study. EBioMedicine. 2024;99 doi: 10.1016/j.ebiom.2023.104930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Behera M.P., Sarangi A., Mishra D., Sarangi S.K. A hybrid machine learning algorithm for heart and liver disease prediction using modified particle swarm optimization with support vector machine. Procedia Comput. Sci. 2023;218:818–827. [Google Scholar]
- 13.Kaur I., Ahmad T. A cluster-based ensemble approach for congenital heart disease prediction. Comput. Methods Progr. Biomed. 2024;243 doi: 10.1016/j.cmpb.2023.107922. [DOI] [PubMed] [Google Scholar]
- 14.Sutarno S., Nurmaini S., Partan R.U., Sapitri A.I., Tutuko B., Rachmatullah M.N., Darmawahyuni A., Firdaus F., Bernolian N., Sulistiyo D. FetalNet: low-light fetal echocardiography enhancement and dense convolutional network classifier for improving heart defect prediction. Inform. Med. Unlocked. 2022;35 [Google Scholar]
- 15.Chowdhury M.M., Ayon R.S., Hossain M.S. An investigation of machine learning algorithms and data augmentation techniques for diabetes diagnosis using class imbalanced BRFSS dataset. Healthcare Analytics. 2024;5 [Google Scholar]
- 16.Griffeth E.M., Stephens E.H., Dearani J.A., Shreve J.T., O'Sullivan D., Egbe A.C., Connolly H.M., Todd A., Burchill L.J. Impact of heart failure on reoperation in adult congenital heart disease: an innovative machine learning model. J. Thorac. Cardiovasc. Surg. 2023;167(6):2215–2225. doi: 10.1016/j.jtcvs.2023.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi H., Yang D., Tang K., Hu C., Li L., Zhang L., Gong T., Cui Y. Explainable machine learning model for predicting the occurrence of postoperative malnutrition in children with congenital heart disease. Clin. Nutr. 2022;41(1):202–210. doi: 10.1016/j.clnu.2021.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Junior J.C., Caneo L.F., Turquetto A.L.R., Amato L.P., Arita E.C.T.C., da Silva Fernandes A.M., Trindade E.M., Jatene F.B., Dossou P.-E., Jatene M.B. Predictors of in-ICU length of stay among congenital heart defect patients using artificial intelligence model: a pilot study. Heliyon. 2024;10(4) doi: 10.1016/j.heliyon.2024.e25406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu X., Jia Q., Yuan H., Qiu H., Dong Y., Xie W., Yao Z., Zhang J., Nie Z., Li X. A clinically applicable AI system for diagnosis of congenital heart diseases based on computed tomography images. Med. Image Anal. 2023;90 doi: 10.1016/j.media.2023.102953. [DOI] [PubMed] [Google Scholar]
- 20.Luo D., Zheng X., Yang Z., Li H., Fei H., Zhang C. Machine learning for clustering and postclosure outcome of adult CHD-PAH patients with borderline hemodynamics. J. Heart Lung Transplant. 2023;42(9):1286–1297. doi: 10.1016/j.healun.2023.05.003. [DOI] [PubMed] [Google Scholar]
- 21.He Q., Liu Y., Dou Z., Ma K., Li S. Congenital heart diseases with airway stenosis: a predictive nomogram to risk-stratify patients without airway intervention. BMC Pediatr. 2023;23(1):351. doi: 10.1186/s12887-023-04160-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reddy C.D., Van den Eynde J., Kutty S. Artificial intelligence in perinatal diagnosis and management of congenital heart disease. Semin. Perinatol. 2022;46(4) doi: 10.1016/j.semperi.2022.151588. [DOI] [PubMed] [Google Scholar]
- 23.Cao H., Wei X., Guo X., Song C., Luo Y., Cui Y., Hu X., Zhang Y. Screening high-risk clusters for developing birth defects in mothers in Shanxi Province, China: application of latent class cluster analysis. BMC Pregnancy Childbirth. 2015;15:1–8. doi: 10.1186/s12884-015-0783-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo Y., Li Z., Guo H., Cao H., Song C., Guo X., Zhang Y. Predicting congenital heart defects: a comparison of three data mining methods. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0177811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacQueen J. Classification and analysis of multivariate observations. 5th Berkeley Symp. Math. Statist. Probability. 1967:281–297. [Google Scholar]
- 26.Alsubaei F.S., Hamed A.Y., Hassan M.R., Mohery M., Elnahary M.K. Machine learning approach to optimal task scheduling in cloud communication. Alex. Eng. J. 2024;89:1–30. [Google Scholar]
- 27.Bezdek J.C., Ehrlich R., Full W. FCM: the fuzzy c-means clustering algorithm. Comput. Geosci. 1984;10(2–3):191–203. [Google Scholar]
- 28.Hussein S. Automatic layer segmentation in H&E images of mice skin based on colour deconvolution and fuzzy C-mean clustering. Inform. Med. Unlocked. 2021;25 [Google Scholar]
- 29.Davies D.L., Bouldin D.W. A cluster separation measure. IEEE Trans. Pattern Anal. Mach. Intell. 1979;(2):224–227. [PubMed] [Google Scholar]
- 30.Song Y., Song W., Yu X., Afgan M.S., Liu J., Gu W., Hou Z., Wang Z., Li Z., Yan G. Improvement of sample discrimination using laser-induced breakdown spectroscopy with multiple-setting spectra. Anal. Chim. Acta. 2021;1184 doi: 10.1016/j.aca.2021.339053. [DOI] [PubMed] [Google Scholar]
- 31.Rousseeuw P.J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987;20:53–65. [Google Scholar]
- 32.Subbalakshmi C., Krishna G.R., Rao S.K.M., Rao P.V. A method to find optimum number of clusters based on fuzzy silhouette on dynamic data set. Procedia Comput. Sci. 2015;46:346–353. [Google Scholar]
- 33.Yazdani M., Zarate P., Kazimieras Zavadskas E., Turskis Z. A combined compromise solution (CoCoSo) method for multi-criteria decision-making problems. Manag. Decis. 2019;57(9):2501–2519. [Google Scholar]
- 34.Liu X.-Y., Wu J., Zhou Z.-H. Exploratory undersampling for class-imbalance learning. IEEE Transactions on Systems, Man, and Cybernetics, Part B (Cybernetics) 2008;39(2):539–550. doi: 10.1109/TSMCB.2008.2007853. [DOI] [PubMed] [Google Scholar]
- 35.Chen C., Liaw A., Breiman L. Using random forest to learn imbalanced data. University of California. 2004;110(1–12):24. Berkeley. [Google Scholar]
- 36.Hido S., Kashima H., Takahashi Y. Roughly balanced bagging for imbalanced data. Stat. Anal. Data Min.: The ASA Data Science Journal. 2009;2(5‐6):412–426. [Google Scholar]
- 37.Seiffert C., Khoshgoftaar T.M., Van Hulse J., Napolitano A. RUSBoost: a hybrid approach to alleviating class imbalance. IEEE Trans. Syst. Man Cybern. Syst. Hum. 2009;40(1):185–197. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sharing research data helps other researchers evaluate your findings, build on your work and to increase trust in your article. We encourage all our authors to make as much of their data publicly available as reasonably possible. Please note that your response to the following questions regarding the public data availability and the reasons for potentially not making data available will be available alongside your article upon publication.
Has data associated with your study been deposited into a publicly available repository?
-
•
Yes
Sharing research data helps other researchers evaluate your findings, build on your work and to increase trust in your article. We encourage all our authors to make as much of their data publicly available as reasonably possible. Please note that your response to the following questions regarding the public data availability and the reasons for potentially not making data available will be available alongside your article upon publication.
Has data associated with your study been deposited into a publicly available repository?
-
•
The data referenced in this manuscript are publicly available and can be accessed through [24]. The dataset utilized in this study comprises non-clinical data spanning six counties in China from 2006 to 2008. It consists of 36,300 instances and encompasses 10 distinct features. Also, we provide the dataset in the GitHub: https://github.com/Amir27Salehi/Congenital-Heart-Disease-