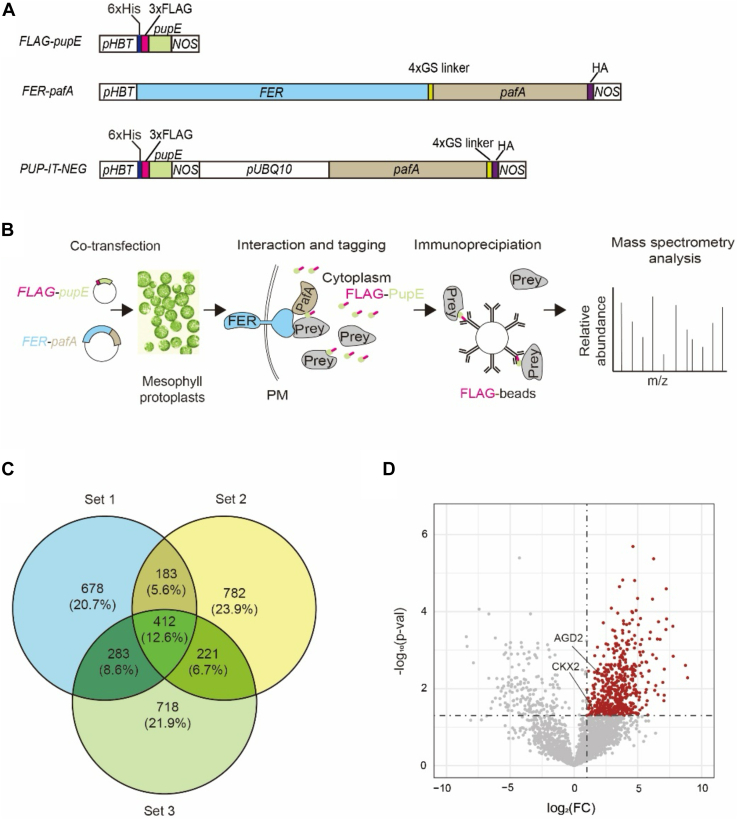
Figure 1.
Identifying FER-proximal proteins by PUP-IT analyses in Arabidopsis protoplasts.A, Gene constructs for transient expression in mesophyll protoplasts. Transcriptional promoter (pHBT) and termination (NOS) sequences respectively flank start and stop codons. FLAG-pupE includes coding sequence for six consecutive histidines (6xHis), three consecutive FLAG epitopes (FLAG) and the activated intermediate of Corynebacterium glutamicum Pup (pupE). FER-pafA includes coding sequence for Arabidopsis FER (FER), four repeats of the amino acid sequence Gly-Gly-Gly-Ser (GS linker), C. glutamicum PafA ligase (pafA) and two consecutive HA epitopes (HA). The PUP-IT-NEG construct includes two genes, the first of which is FLAG-pupE. The second is flanked by the promoter of Arabidopsis UBQ10 (pUBQ10) and NOS and includes coding sequence of pafA, GS linker and HA. B, Schematic diagram of PUP-IT for FER-interaction tagging in Arabidopsis protoplasts. Transfection introduced FLAG-pupE and FER-PafA to mesophyll protoplasts. Interaction of prey proteins and plasma membrane (PM)-localized FER-PafA led to ligation of FLAG-PupE to prey proteins. Immunoprecipitation with anti-FLAG antibody-beads enriched FLAG-tagged prey proteins. Analysis of peptides from prey proteins using mass spectrometry identified candidate proteins. C, overlap of three sets of candidate proteins from three replicated experiments is depicted in a Venn diagram with numbers and percentages of proteins. D, Volcano plot of the 2464 (red and grey) proteins identified by LC–MS/MS in protoplasts. The −log10 (p-value) is plotted against the log2 (fold change: abundant proteins in the experiment group/abundant proteins in the control group). The non-axial vertical lines denote ±1.0-fold change while the non-axial horizontal line denotes p = 0.05. The red dots represent 491 proteins with significant differences between the experimental group and the control (p-value <0.05 and fold change >1).