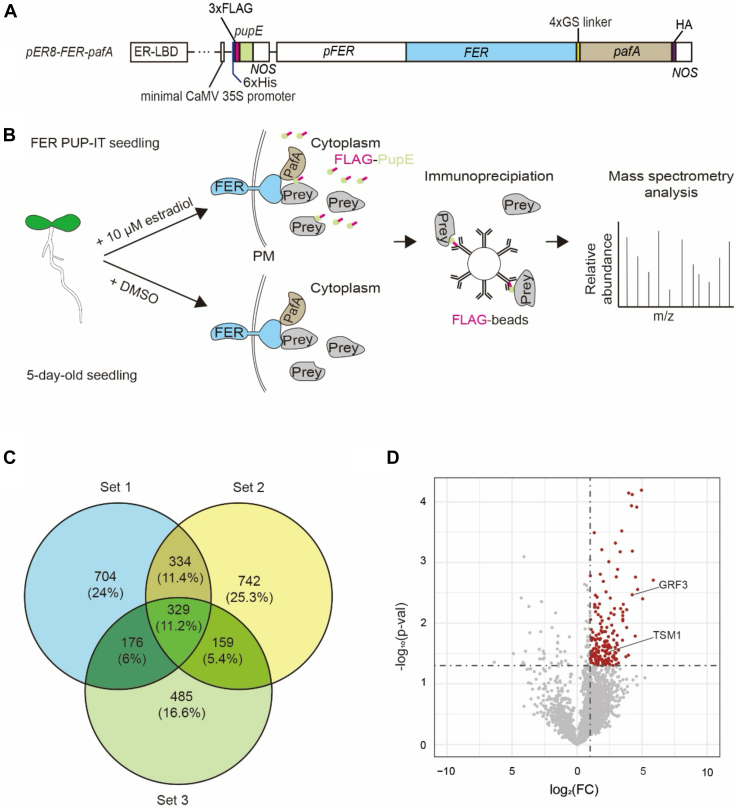
Figure 2.
Identifying FER-proximal proteins by PUP-IT analyses in Arabidopsis seedlings.A, transfer DNA (T-DNA) region of the binary plasmid used for Agrobacterium-mediated transformation of Arabidopsis includes three genes XVE, 8xLexO-FLAG-pupE and pFER-FER-pafA. XVE constitutively expresses an estrogen receptor-based transactivator (27). The promoter sequence 8xLexO makes transcriptional expression of FLAG-pupE (described in Fig. 1A) responsive to estradiol-activated XVE (27). The promoter sequence of FER (pFER) gives endogenous expression to FER-pafA (described in Fig. 1A). B, Schematic Diagram of PUP-IT for FER-interaction tagging in Arabidopsis seedlings. Five-day-old FER PUP-IT transgenic seedlings were treated for 2 days with estradiol to induce FLAG-pupE or with diluted DMSO alone to leave expression uninduced. PM-localized FER-PafA ligated FLAG-PupE to prey proteins. FLAG-tagged prey proteins were immunoprecipitated on anti-FLAG antibody-beads. Mass spectrometric analysis was used to identify peptides from enriched prey proteins. C, the overlap of three sets of candidate proteins from three replicated experiments is depicted in a Venn diagram with numbers and percentages of proteins. D, Volcano plot of the 1964 proteins (red and grey) identified by LC–MS/MS in the seedling. The −log10 (p-value) is plotted against log2 (fold change: abundant proteins in the experiment group/abundant proteins in the control group). The non-axial vertical lines denote ±1.0-fold change while the non-axial horizontal line denotes p = 0.05. The red dots represent 182 proteins with significant differences between the experimental group and the control (p-value <0.05 and fold change >1).