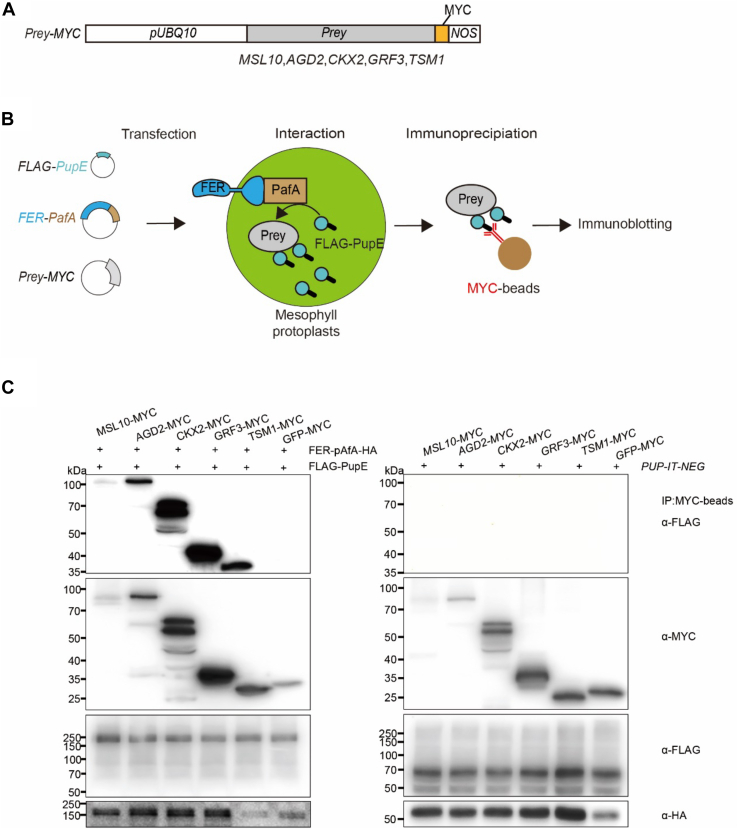
Figure 4.
Validation of candidate FER-Interacting Proteins in Arabidopsis protoplasts.A, Prey-MYC constructs for transient expression of candidate prey proteins in protoplasts. The Arabidopsis UBQ10 promoter (pUBQ10) and NOS sequences respectively flank start and stop codons. Constructs include coding sequences of MSL10, AGD2, CKX2, GFR3, TSM1, or GFP (negative control) and two copies of the MYC epitope. B, Schematic diagram of PUP-IT for validating FER-interacting candidate prey proteins in mesophyll protoplasts. Prey-MYC constructs were transiently co-expressed with FLAG-pup and FER-pafA overnight in transfected protoplasts, FLAG-Pup tags on Prey-MYC captured interactions with FER-pafA. Prey-MYC proteins were immunoprecipitated (IP) with anti-MYC antibodies immobilized on agarose beads (MYC-beads), separated in SDS-PAGE and immunoblotted with antibodies to FLAG (α-FLAG), HA (α-HA) and MYC (α-cMYC) epitopes. C, validating FER-interacting candidate prey proteins. α-cMYC detected bands with gel mobilities unique to expressed Prey-MYC proteins. MSL10 (83 kDa), AGD2 (88 kDa), CKX2 (56 kDa), GRF3 (33 kDa), TSM1 (26 kDa) and GFP (28 kDa); and, α-HA detected a band with gel mobility expected for FER-PafA-HA (155 kDa) and PafA-HA (58 kDa). On immunoblots of IP on MYC-beads, α-FLAG detected bands with gel mobility unique to expressed Prey-MYC proteins (right) and failed to detect bands for the negative control (left) (from expression of GFP-MYC or PUP-IT NEG). kDa, kilodaltons.