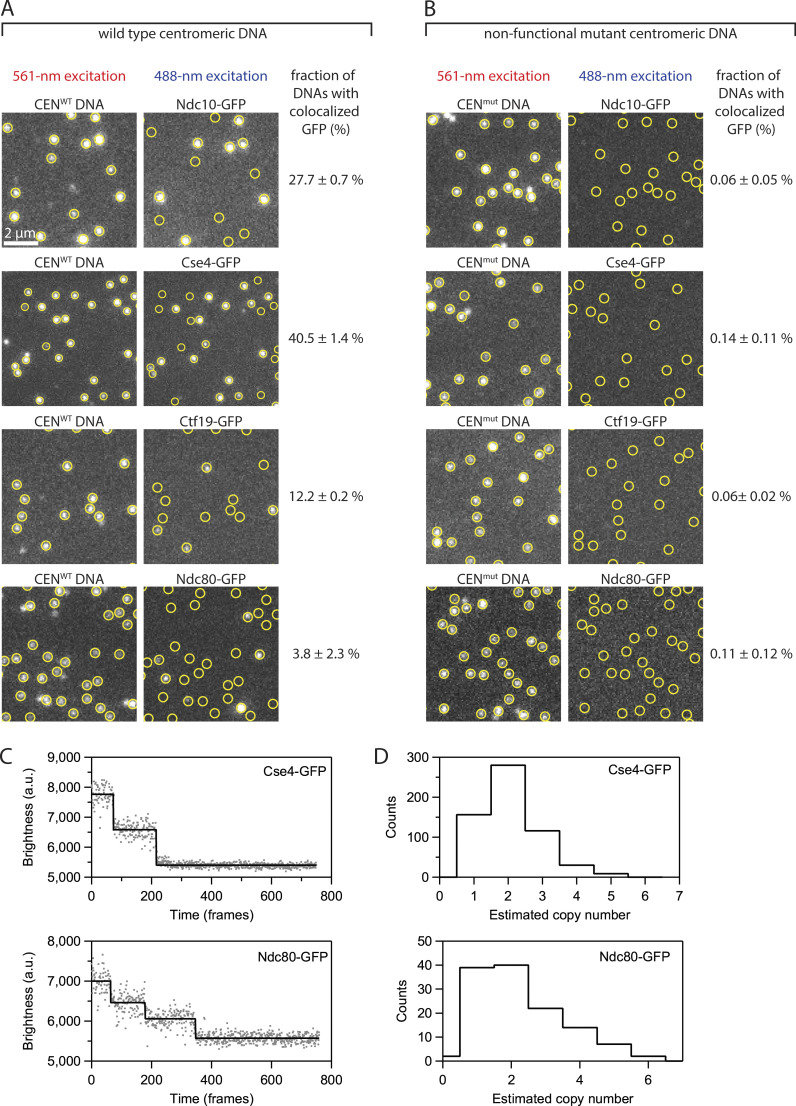
Figure S1.
De novo assembly of individual kinetochores occurs specifically on centromeric DNAs. (A) Images of individual Atto565-labeled centromeric DNAs (left) and corresponding images of GFP-tagged kinetochore subcomplexes (right) from the same fields of view. Yellow circles mark locations of individual wild type centromeric DNAs (CENWT), which contain the complete 117-bp centromere sequence from S. cerevisiae chromosome III. (B) Images from negative control experiments using mutant centromeric DNAs (CENmut) carrying a 3-bp substitution that prevents kinetochore assembly. The percentages in both A and B represent average fractions (±SEM) of centromeric DNAs that colocalized with a GFP signal from the indicated kinetochore protein, calculated from N > 3,400 DNAs for each kinetochore component from at least nine fields of view across three independent experiments. (C) Photobleach analysis of Cse4- or Ndc80-GFP assembled kinetochores. Representative records of fluorescence intensity versus time for individual Cse4-GFP (top) or Ndc80-GFP (bottom) assembled kinetochores. The raw intensity data is represented by gray spots and the estimated bleach steps are represented by the solid black line. Bleach steps were estimated using the Tdetector2 step detection algorithm (Chen et al., 2014). (D) Histograms showing the estimated copy number of Cse4-GFP (top) or Ndc80-GFP (bottom) present in individually assembled kinetochores. N = 591 individual Cse4-GFP kinetochore assemblies and N = 126 individual Ndc80-GFP kinetochore assemblies.