Extended data Fig. 7 ∣. Generation of COBLL1 mutant mice using CRISPR/Cas9 editing.
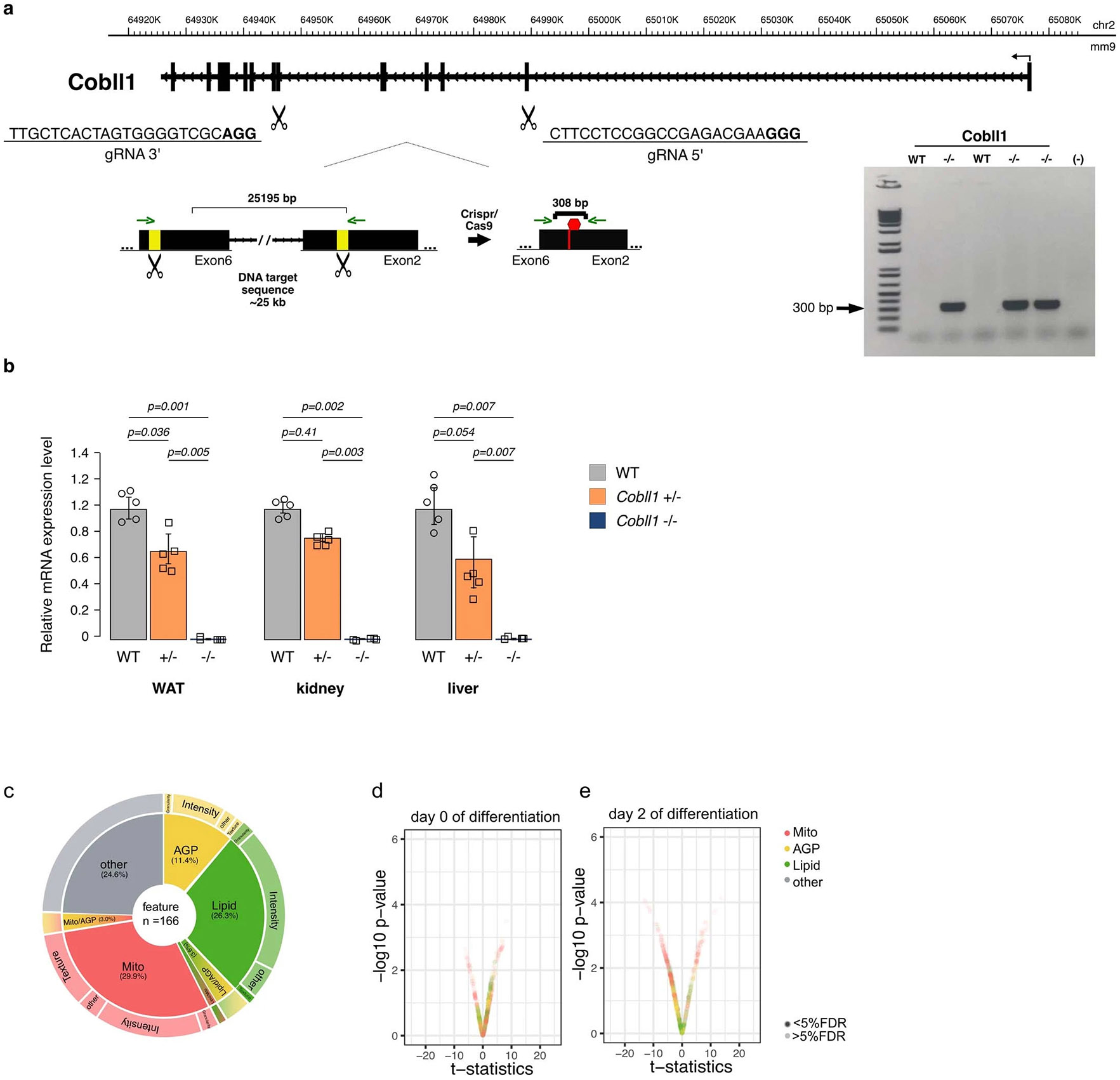
(a) Overview of the CRISPR/− Cas9 strategy to delete ~20 kb of the Cobll1 gene. The gRNA-targeting sequences (gRNAs) are underlined, and the PAM sequences are indicated in bold. Exons are represented as thick black boxes introns are indicated as black lines with arrows, and the yellow boxes indicate the DNA-targeting region. Red hexagon indicates a stop codon gen- erating a Cobll1 truncated protein. Agarose gel showing the PCR products generated from DNA containing success- fully targeted Cobll1 from F0 mouse tail genomic DNA. The 308 bp band corresponds to the genomic deletion. (b) A real-time quantitative PCR of levels of Cobll1 mRNA in white adipose tissue (WAT), liver and kidney of Cobll1 WT, Cobll1 heterozygous (+/−) and null knockout Cobll1 (−/−) animals to confirm the Cobll1 ablation in knockout animals. Each group was analyzed using 5 different mice and the values were expressed as the mean ± s.e.m and P values by Student’s t-test the experiment was repeated independently two times with similar results. (c) Pie chart illustrating non-redundant differential features per channel and class of measurement at day 8 of subcutaneous adipocyte differentiation in rs6712003 homozygous risk compared to non-risk carriers. (d, e) Differences in morphological profiles between AMSCs from Cobll1−/− mice (n = 3) and WT (n = 4) at (h) day 0 (i) day 2 (t-test two-sided, significance level < 5%FDR).
