Extended data Fig. 3 ∣. Chromatin inter- actions and CRISPRi of 2q24.3 locus identify COBLL1 as target genes.
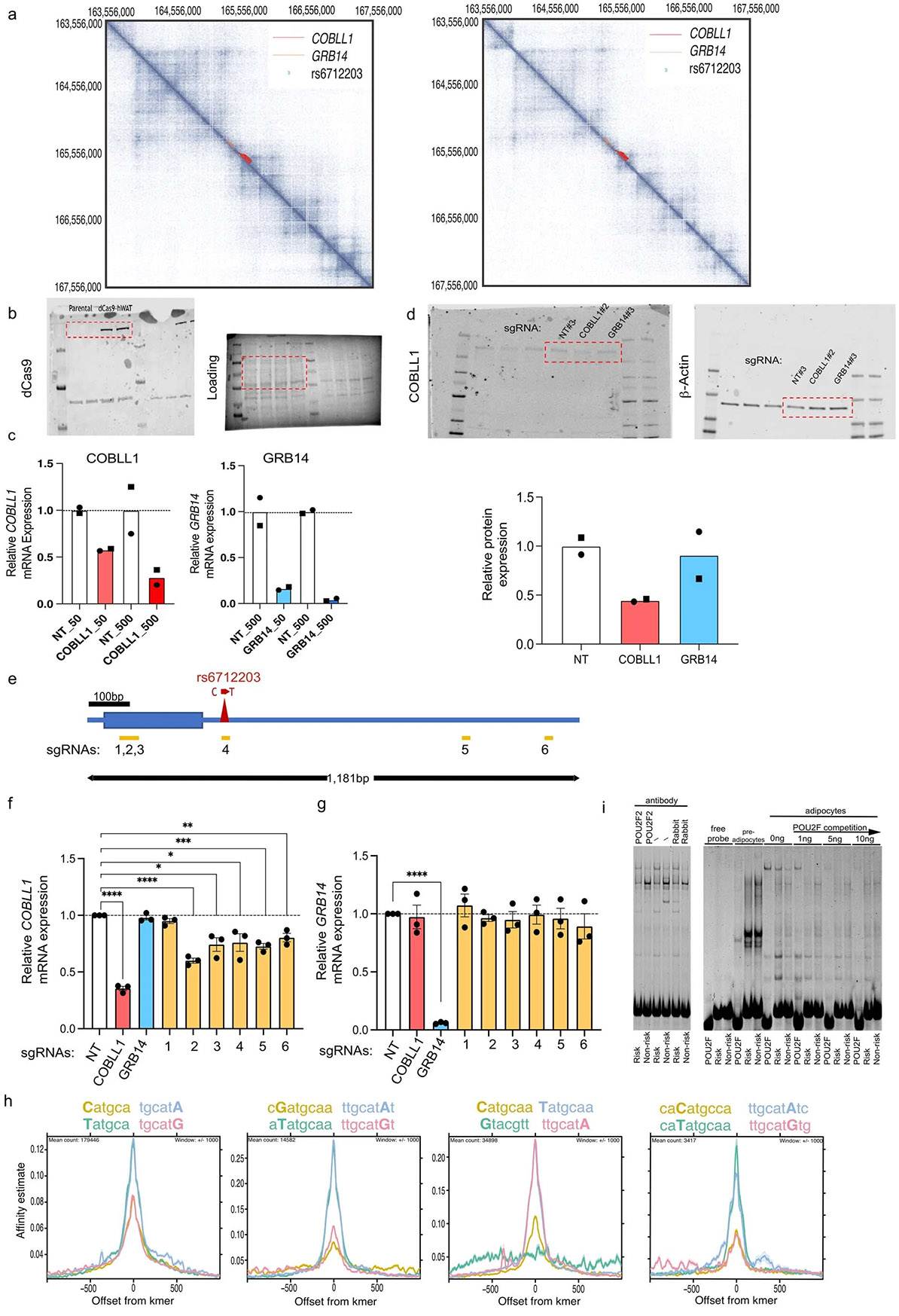
(a) Cross-cell type conserved genome-wide higher order chromatin interactions for the 2q24.3 locus analyzed by Hi-C assays in human fibroblasts (left) and NHEK primary normal human epidermal keratinocytes (right), chr2: 163,556,000 - 167,558,000 (hg19), binned at 2 kb resolution. (b) Cas9 protein expression in dCas9 hWAT compared to the parental hWAT cell line. (c) mRNA expression of COBLL1 and GRB14 in response to increasing amounts of lentiviral sgRNA vectors (2 sgRNAs, virus volume 50 μl and 500 μl) targeting TSS regions of each gene compared to non-targeting controls (NT, 2 sgRNAs). Columns are means of individual sgRNAs indicated by different symbols. (d) COBLL1 protein expression normalized to b-actin in dCas9 hWATs transduced with sgRNAs targeting COBLL1 or GRB14 compared to controls. Top panel: Image of gel of representative sgRNA targeting NT, COBLL1 or GRB14. Bottom panel: plot of protein expression; 2 sgRNA for each target in 2 replicates. (e) Representation of 1,181 bp region flanking the COBLL1 intronic variant rs6712203 at the 2q24.3 MONW locus showing individual sgRNAs (n = 6) targeting the rs6712203 flanking regulatory region used in the CRISPRi experiments. (f, g) mRNA expression of (f) COBLL1 and (g) GRB14 in undifferentiated dCas9-hWAT preadipocytes at 6 days post lentiviral transduction with sgRNAs targeting TSS regions (red: COBLL1 TSS; blue GRB14 TSS) and the rs6712203-flanking regulatory element at position 1 to 6 as depicted in (e). Data are mean +/− SEM of 3 independent experiments. **** P < 0.0001, *** P = 0.0004, ** P = 0.006, * P = 0.013 – 0.036, two-tailed Student’s t test. (h) Predicted binding of POU2F2 between the two alleles using the Intragenomic Replicate Method (Cowper-Sal lari et al. 2012). As in Fig. 2d with different kmer counts.
