Abstract
In the brain, AMPA-type glutamate receptors (AMPARs) form complexes with their auxiliary subunits and mediate the majority of fast excitatory neurotransmission. Signals transduced by these complexes are critical for synaptic plasticity, learning and memory. The two major categories of AMPAR auxiliary subunits are TARPs and cornichon homologues (CNIHs), which share little homology and play distinct roles in controlling ion channel gating and trafficking of AMPAR. Here, I report high-resolution cryo-EM structures of AMPAR in complex with CNIH3. Contrary to its predicted membrane topology, CNIH3 lacks an extracellular domain and instead contains four membrane-spanning helices. The structure shows the interaction interface that dictates channel modulation and reveals lipids surrounding the complex. These structures provide insights into the molecular mechanism for ion channel modulation and assembly of AMPAR/CNIH3 complexes.
One Sentence Summary:
High-resolution cryo-EM reveals how an auxiliary subunit interacts with an ion channel to regulates synaptic function.
AMPA-type ionotropic glutamate receptors (AMPARs), ligand-gated ion channels activated by the neurotransmitter glutamate, mediate the majority of fast excitatory synaptic transmission in the brain (1). They regulate synaptic plasticity, which in turn influences circuit activity, cognition, learning, and behavior. Their dysfunction is associated with a variety of neurological and psychiatric disorders (2), including major depression disorder, Alzheimer’s disease, Rasmussen’s and limbic encephalitis, seizure, cognitive dysfunction, and autism.
In mammals, the pore forming subunits of AMPARs are called GluA1 to 4. The extracellular domains of each subunit consist of an amino terminal domain (NTD) and a ligand binding domain (LBD) (Fig 1A). In the resting state, NTDs are stacked on top of the LBD and extend away from the membrane (3). Glutamate binds to the LBD and induces a conformational change in the transmembrane domain (TMD), that in turn, triggers gating (4). The TMD forms the ion channel pore made of three membrane-spanning segments (M1, M3, and M4) and a reentrant element (M2). A short cytoplasmic domain (CTD) extends into the cytoplasm. AMPARs function as tetramers and adopt a dimer-of-dimers architecture resembling a ‘Y’ shape, in which the extracellular NTD and LBD each form dimers exhibiting an overall 2-fold symmetry, while the membrane embedded portion adopts a near 4-fold symmetry (5) (See organization of GluA2 tetramer in Fig 1C). In addition, certain heterotetrameric AMPAR adopts a compact global conformation that differ from the canonical ‘Y’-shape (6).
Fig. 1. Cryo-EM structures of the complex formed of GluA2 and CNIH3 (A2-C3).
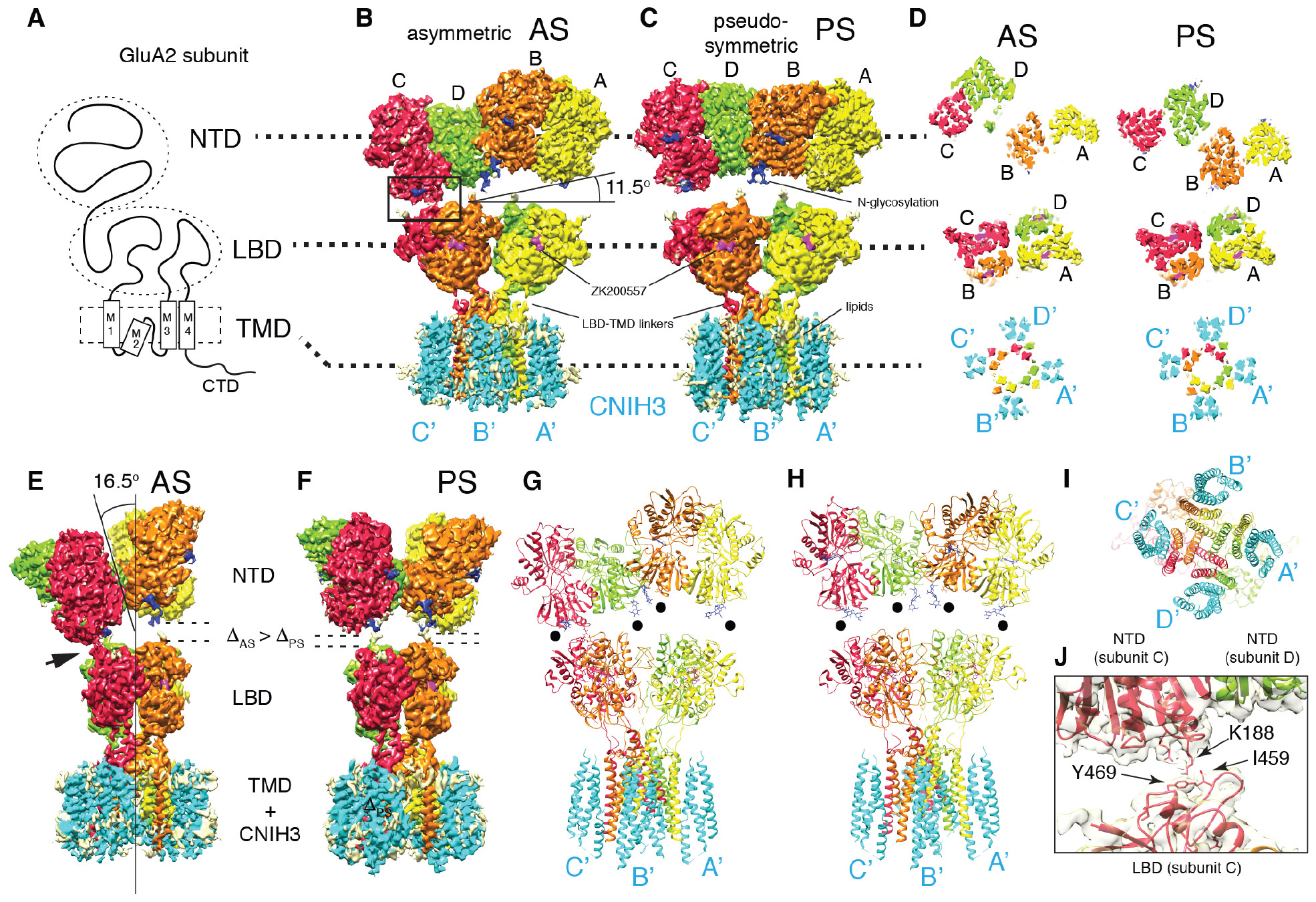
A. Domain organization of a GluA2 subunit.
B-C. Density map of A2-C3 in AS and PS. From this view the NTD layer is tilted by 11.5° in AS. No symmetry was imposed in solving AS, whereas C2 was imposed for PS. Visualizing thresholds: NTD(AS) at 7.07σ, TMD-LBD-C3(AS) I at 7.56σ, NTD(PS) at 7.32σ, TMD-LBD-C3(PS) at 6.80σ. Overall resolutions: NTD(AS)=3.1Å, LBD-TMD-C3(AS) I=3.5Å, NTD(PS)=3.1Å, LBD-TMD-C3=3.2Å. See TableS1 and FigS3-8 for detailed description of each map.
D. Cross sections of each domain indicated by dashed lines, viewed from the top (the NTD side). The subunits of tetrameric GluA2 are referred to as A (yellow), B (orange), C (red), and D (green). The cyan densities are CNIH3, named A’-D’ based on location, following the style used for TARPs (16, 25).
E-F. Side views of maps shown in B and C. The tilt angle of the NTD layer (16.5°), the NTD-LBD contact (arrow head), gaps between NTD and LBD (ΔAS, ΔPS) are indicated.
G-I. Molecular models of A2-C3 in AS and PS are shown as ribbon diagrams. Models were built from maps NTD(AS), LBD-TMD-C3(AS)II, NTD(PS), and LBD-TMD-C3(PS) (TableS1). Black dots indicate glycosylation at N241. Bottom view I.
J. Zoomed in view of the NTD-LBD contact in C subunit indicated as a rectangle in B. Model and map (NTD(AS) at 7.07σ) are superimposed.
AMPARs form complexes with various structurally unrelated auxiliary subunits, which are membrane proteins that regulate AMPAR trafficking or gating (and in some cases both) (7). The two major classes of AMPAR auxiliary subunits belong to the claudin homolog and cornichon homolog (CNIHs), which share little homology (8). TARPs are members of the claudin homolog family and are the most extensively studied (7-9). Among the cornichon family, CNIH2 and 3 (CNIH2/3) are AMPAR auxiliary subunits (10). Unlike the TARPs, CNIH2/3 function at the endoplasmic reticulum where, in mammals, they may control assembly of heteromeric AMPARs (11, 12). CNIH2/3 remain associated with the synaptic AMPAR complex and, in many cases, co-assemble with TARPs (11, 13, 14). Both TARPs and CNIH2/3 slow ion channel desensitization to varying degrees (7). The role of auxiliary subunits in tuning ion channel gating kinetics is predicted to have significant impact on circuit dynamics (7). Knowledge on the modulation mechanisms of AMPAR gating and trafficking used by various auxiliary subunit could guide rational design of new therapeutic compounds. Currently, our structural knowledge of AMPAR auxiliary subunit complexes has been limited to those that contain either TARPs or GSG1L, which are both claudin homologues (15, 16).
I investigated cryo-EM structures of complexes composed of GluA2 and CNIH3 (hereafter referred to as A2-C3) bound to an antagonist ZK200775 (280μM). Detailed methods on sample preparation and data collection are given in the Supplementary Materials. In the first step of analyses, I obtained full-length structures at overall resolution of 4.4Å (Fig S3-4). The small flexibility between NTD and LBD interfered with further high-resolution analysis, and thus in the second step the NTD layer and the rest of the complex (hereafter referred to as LBD-TMD-C3) were analyzed separately by focused classification and refinement (Extended text, FigS3-10). High-resolution maps, whose overall resolutions ranged between 3.0 to 3.5Å, and their molecular models (for statistics see TableS1) were placed into the 4.4Å resolution full-length map to reconstruct the complete structure (Fig1).
I observed two global conformations. The pseudo-symmetric conformation (PS) resembles the canonical ‘Y’ shape (Fig1C), whereas the asymmetric conformation (AS) exhibits tilted NTD layer relative to the LBD layer, making inter layer contact at one corner (Fig1B-D). Similar numbers of particles (AS=218,413 and PS=190,470) contributed to each conformation. The maps of PS and AS revealed four extra densities in the micelle attached to the TMD of GluA2, which are CNIH3 bound at a stoichiometry of GluA2 : CNIH3 = 4 : 4 (Fig1B-C).
The architectures of NTD tetramer were virtually identical between AS and PS (RMSD of Cα = 0.374Å), and similar to previous reports (FigS11). Glycosylation at N241, which was eliminated by mutation in previous studies (5, 17), points towards LBD, potentially biasing the NTD layer tilted in AS and preventing it from descending vertically (Fig1B-C, F-G). In AS, K188 is close to I459 and Y469 at the inter domain contact between NTD and LBD (Fig1B, J). The NTDs might alter dynamics of LBDs through direct interaction, which could potentially result in allosteric gating modulation, such as seen in NMDA receptors (18). The LBDs are nearly identical between AS and PS (RMSD of Cα = 0.542Å), and also similar to those of GluA2/stargazin and GluA2/GSG1L in the closed state (15, 19) (FigS12). Consistently, architectures below the NTD layer were virtually identical between AS and PS (RMSD of Cα = 0.526Å, FigS7E). Switching between AS and PS must be a probabilistic process, requiring the native NTD-LBD linker. The pore is closed and the lower part of the channel is more compacted compared to GluA2 tetramer with no auxiliary subunit (PDB:3KG2). This compaction geometrically occludes the M2, which was unresolved (FigS13) (see Extended Text).
An atomic model of CNIH3 was built de novo and produced the first molecular view of a member of the cornichon family. Previous studies have relied on computational models that predict CNIHs having their N-termini in the cytoplasm and spanning the membrane only three times (8, 10, 20-22). Our data, however, redefines the topology of CNIH3 and reveals a geometry resembling four transmembrane segments with extracellular N- and C-terminus (Fig2A-B). CNIH3 lacks a canonical signal peptide but the amino terminus, which remains uncleaved and buried in the membrane, appears to substitute for it. The first 12 amino acids are almost identical in mammals, flies, and worms, but not in plants or yeast (Fig2C). I will refer to this fragment as uncleavable membrane inserting peptide (UMIP). UMIP of CNIH2 is intolerant to missense mutations in humans (23)(FigS14).
Fig. 2. Membrane topology of CNIH3.
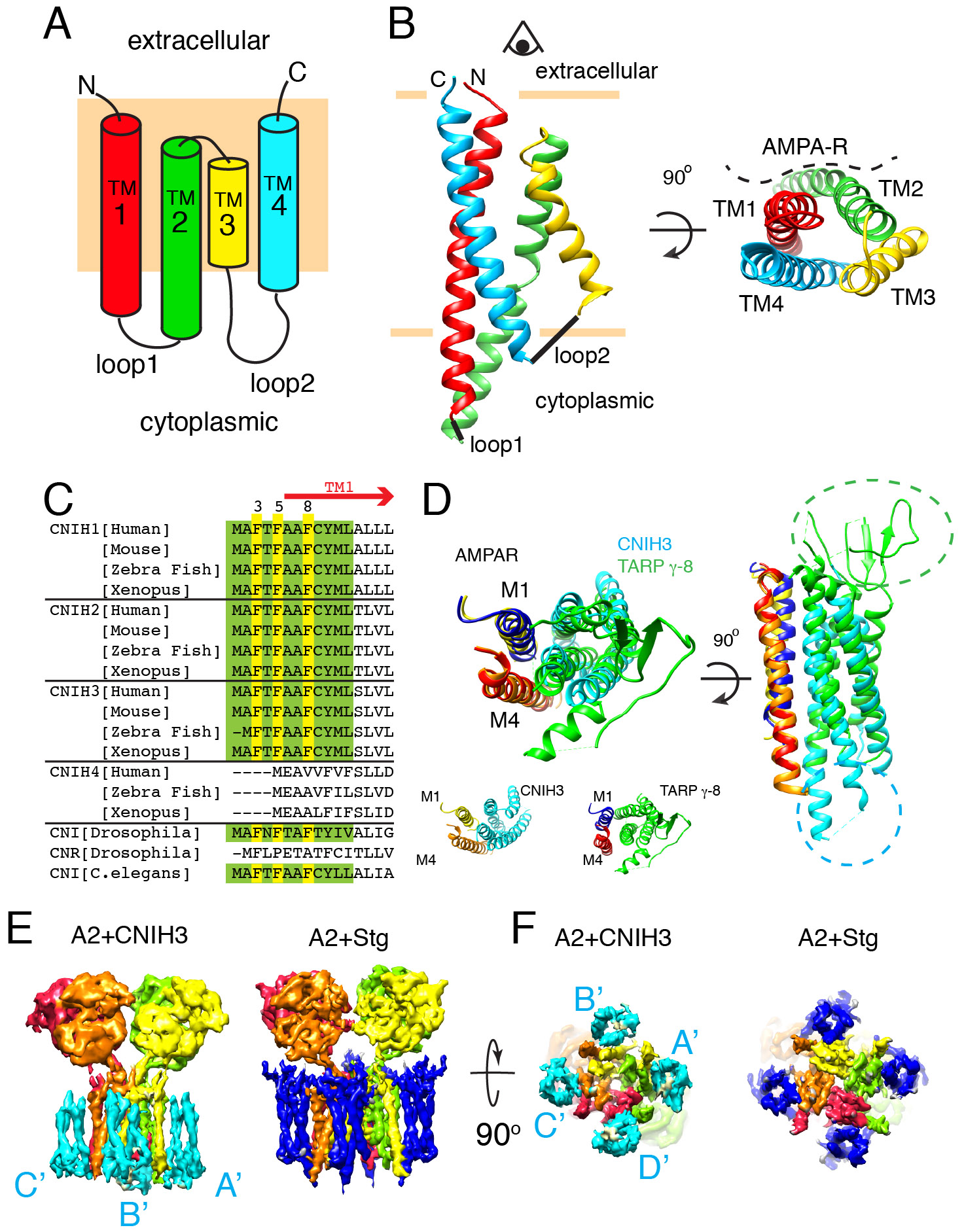
A. Schematic of the topology. Rectangle (pale orange) in the background represents the membrane.
B. Ribbon diagram of CNIH3 shown from side (left) and top (right) views. The helices are colored according to the topology diagram in A. The location of AMPAR is shown in the top view. The model was built from map LBD-TMD-C2 (TableS1).
C. Sequence alignment of the N-terminal fragment that contains the UMIP (green, and yellow). F3, 5, and 8 (yellow) play critical role in complex assembly (see Fig3E).
D. The M1 and M4 of adjacent subunits of AMPAR form the binding surface for both CNIH3 and TARP. Aligning M4 helix is sufficient to superimpose remaining helices (M1, and TM1-4 of CNIH3 or TARP γ-8). The models before alignment are shown on the bottom. The side view reveals the unique extracellular and cytoplasmic extensions of TARP γ-8 (green) and CNIH3 (cyan).
E and F. Density map of GluA2-CNIH3 complex in PS (map A2-C3(PS) at 6.91σ) is compared with that of GluA2-stargazin complex (EMDB-8721 at 25.1σ) at a comparable resolution. NTD is excluded from display. CNIH3 and stargazin are shown in cyan and blue, respectively. Side and bottom views are shown.
The TM1 helix begins within UMIP and the end of the helix penetrates into the cytoplasm. After a short unresolved loop the TM2 starts as a cytoplasmic helix, which was misinterpreted as an extracellular loop in previous studies (8, 10, 20-22). Interestingly, TM2 turns 180 degree in the membrane and connects to TM3 that reenters the cytoplasm. A tryptophan (W88) at the junction between TM2 and 3 is conserved among cornichons (FigS15). The folded-jack-knife shape of TM2 and 3 may be a signature of all cornichons. The majority of CNIH3 is embedded in the membrane with a small cytoplasmic domain, and thus a direct interaction between the LBD of GluA2 and CNIH3 in the extracellular space is unlikely. In contrast, TARPs and GSG1L modulates AMPARs by directly contacting the LBD in the extracellular space (24, 25). Based on the structure and the locations of functionally significant mutations of CNIH3, receptor modulation must occur via the intra-membrane and the cytoplasmic interaction between the two proteins (21, 26) (see Extended text).
CNIH3 binds to the M1 and M4 of adjacent subunits of GluA2 where TARPs and GSG1L associate (Fig2D) (15, 16). Despite absence of homology, the bundle of four helices of CNIH3 resembles the geometry of TARPs and GSG1L. Geometric conservation extends to the M1 and M4 of GluA2 that interface with the auxiliary subunits. Indeed, the helices of CNIH3 and TARP γ-8 together with the M1 and 4 of GluA2 can be superimposed, when the alpha carbon backbone of M4 of both complexes are aligned at RMSD=0.763Å (Fig2D). The detailed residue contacts are different (see Extended Text, FigS16), even though the overall architectures of the complexes appear similar at low resolution (Fig2E-F).
Numerous non-protein densities (L-a – L-h), whose features are characteristic of either lipids or detergents, surround the hydrophobic surface (Fig3A-B). These densities were best resolved in the map LBD-TMD-C3lipid, which was calculated from a smaller number of particles than what was used to generate the higher resolution map of LBD-TMD-C3 (see Methods and FigS9), indicative of a small structural variability in the lipids and detergents that are associated with the complex. L-a to L-e and L-h were visible at a wide range of thresholds and modeled as acyl groups. L-h is a component of the inner (cytoplasmic) leaflet of the lipid bilayer and occupies the space where M2 is typically located, making contacts with both GluA2 and CNIH3 (FigS17). The tips of L-c and L-h contact each other near the center of the membrane (Fig3D). L-c, an outer leaflet component, contacts both M1 (Y523 and V530) and M3 (F607) of adjacent subunits (Fig3C and S17), and approaches the center of the ion channel similar to the lipid density L4 found in the heteromeric AMPAR in complex with TARP γ-8 (25). However, the contact points of L-c and L4 with AMPAR were different, except for F607 of M3. I hypothesize that the contrasting lipid geometry of AMPAR in complex with TARP γ-8 and CNIH3 contributes to their functional differences.
Fig. 3. Binding interface and arrangements of lipids.
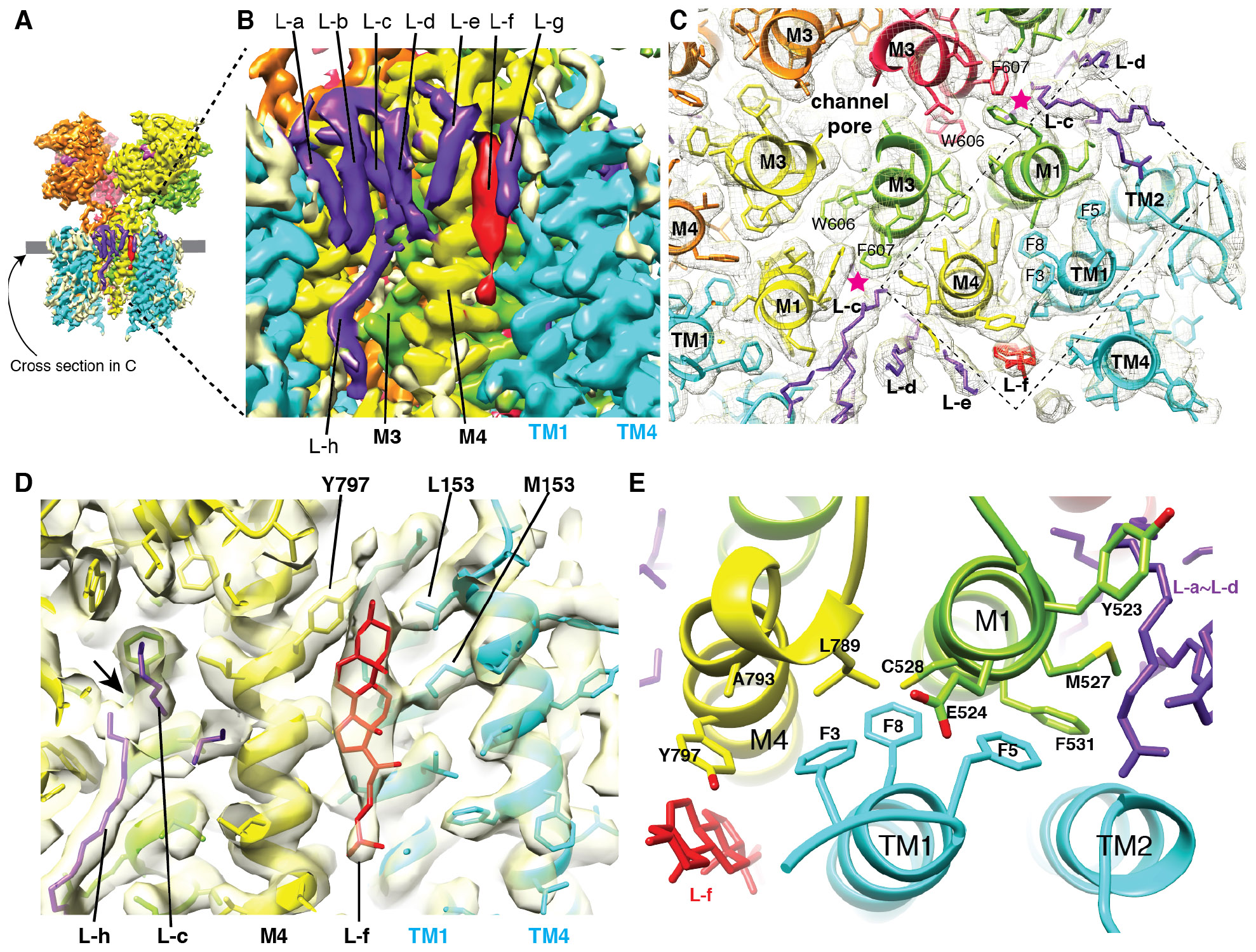
A. The densities that surround the transmembrane helices have characteristic appearance of densities derived from lipid and detergents. The map LBD-TMD-C3lipid (see Methods and FigS9) is displayed at 6.02σ.
B. A magnified view of the area in A where the lipid-like densities are attached to the complex (L-a – L-h). The tips of the L-c and L-h make contact. L-f is a bulkier density whose contour resembles a cholesterol group. Visualization threshold set at 6.02σ.
C. Cross section at the level indicated by the gray line in A. The map and model are superimposed. Stars (magenta) indicate contacts between L-c and F607(M3). The details within the dashed rectangle are in E.
D. Superimposed density map and a molecular model, showing contacts made between L-f (cholesterol) and side chains of GluA2, and CNIH3. The tips of the L-c and L-h make contact at the arrow. Visualization threshold set at 5.32σ.
E. Molecular architecture of the GluA2-CNIH3 interface. F3, 5, and 8 of CNIH3 (cyan) contact residues in M1 (E524, M527, C528, and F531) and M4 (L789, A793, and Y797) of adjacent subunits of GluA2. Y797 (M4) and M527 (M1) simultaneously contact lipid-like densities L-f and L-b, respectively. Y523 (M1), which does not contact CNIH3, is immediately next to the interface and contacts L-c.
A bulkier lipid (L-f) in A2-C3, and interpreted as a cholesterol group, sits next to the interaction interface between GluA2 and CNIH3, making contacts with both M4 (Y797) and TM1 (L157 and M153) (Fig3D, S17). Within the interface, three phenylalanines (F3, 5, and 8) of CNIH3 make contacts with M1 (E524, M527, and C528) and M4 (L789, A793, and Y797) of GluA2 (Fig3E). In particular, Y797 interacts with both L-c and CNIH3. Except for Y797, the residues at the interface are unique to AMPARs and replaced by different residues in closely related kainate receptors that dose not interact with CNIH3, establishing specificity for assembly. A previous study has demonstrated that introducing mutations to residues L528, L789, and A793, now shown to be at the interface, destabilize the complex and bi-directionally alter the magnitude of gating modulation by CNIH3 (26). Replacing A793 of M4 and F3 of CNIH3 to cysteine induced disulfide crosslink, further supporting the model that two residues interact (FigS18). I hypothesize that the binding site for the three phenylalanines (F3, 5, and 8 of CNIH3) near the extracellular surface of GluA2 dictates gating modulation and is a potential target for drugs that could be used to control the ion channel activity of AMPARs. The detailed architecture of the interaction interface suggests a possible role for lipids in regulating the assembly or function of the AMPAR-auxiliary subunit complexes. The molecular model of CNIHs is likely to serve as a reference for future investigations on their biology.
Supplementary Material
Acknowledgements:
I thank Elena Zaika for technical support in isolating clone #7. I acknowledge the use of the Molecular Cryo-EM facility (supported by Scott Collier, Elad Binshtein, and Melissa Chambers) and computation resources (ACCRE and DORS) at Vanderbilt University. I thank Caleigh Azumaya, Aichurok Kamalova, Erkan Karakas, Chuck Sanders, Rado Danev, and Ingo Greger for discussion. I thank Masahide Kikkawa and Haruaki Yanagisawa for providing access and facilitating data collection using the Titan Krios at the University of Tokyo. I thank Walter Chazin and Roger Colbran for support. Software was provided by SBgrid.
Funding:
This work was supported by funding from NIH (R01HD061543 to TN, S10RR031634 to Jarrod Smith) and Vanderbilt University (to TN).
Footnotes
Competing interests: The author declares no competing interests.
Data and materials availability: Reagents and other materials will be available upon request to TN. The structural data are deposited as EMDB IDs: 20330, 20332, 20645, 20666, 20717, 20727, 20732, 20733, 20734, and PDB IDs: 6PEQ, 6U5S, 6U6I, 6UCB, 6UD4, 6UD8.
References and Notes:
- 1.Traynelis SF et al. , Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 62, 405–496 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowie D, Ionotropic glutamate receptors & CNS disorders. CNS Neurol Disord Drug Targets 7, 129–143 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T, Structure and different conformational states of native AMPA receptor complexes. Nature 433, 545–549 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Armstrong N, Gouaux E, Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28, 165–181 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Sobolevsky AI, Rosconi MP, Gouaux E, X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745–756 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herguedas B et al. , Structure and organization of heteromeric AMPA-type glutamate receptors. Science 352, aad3873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson C, Nicoll RA, The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits. Neuron 70, 178–199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwenk J et al. , High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron 74, 621–633 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Shanks NF et al. , Differences in AMPA and kainate receptor interactomes facilitate identification of AMPA receptor auxiliary subunit GSG1L. Cell Rep 1, 590–598 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwenk J et al. , Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science 323, 1313–1319 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Herring BE et al. , Cornichon proteins determine the subunit composition of synaptic AMPA receptors. Neuron 77, 1083–1096 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brockie PJ et al. , Cornichons Control ER Export of AMPA Receptors to Regulate Synaptic Excitability. Neuron 80, 129–142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu X et al. , GSG1L suppresses AMPA receptor-mediated synaptic transmission and uniquely modulates AMPA receptor kinetics in hippocampal neurons. Nat Commun 7, 10873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boudkkazi S, Brechet A, Schwenk J, Fakler B, Cornichon2 dictates the time course of excitatory transmission at individual hippocampal synapses. Neuron 82, 848–858 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI, Channel opening and gating mechanism in AMPA-subtype glutamate receptors. Nature, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S et al. , Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM. Cell 170, 1234–1246 e1214 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yelshanskaya MV, Li M, Sobolevsky AI, Structure of an agonist-bound ionotropic glutamate receptor. Science 345, 1070–1074 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tajima N et al. , Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature 534, 63–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, Chen S, Yoshioka C, Baconguis I, Gouaux E, Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM. Nature 536, 108–111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wudick MM et al. , CORNICHON sorting and regulation of GLR channels underlie pollen tube Ca(2+) homeostasis. Science 360, 533–536 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Shanks NF et al. , Molecular Dissection of the Interaction between the AMPA Receptor and Cornichon Homolog-3. J Neurosci 34, 12104–12120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Y et al. , Functional comparison of the effects of TARPs and cornichons on AMPA receptor trafficking and gating. Proc Natl Acad Sci U S A 107, 16315–16319 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Traynelis J et al. , Optimizing genomic medicine in epilepsy through a gene-customized approach to missense variant interpretation. Genome Res 27, 1715–1729 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI, Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes. Neuron 94, 569–580 e565 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herguedas B. et al. , Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP gamma8. Science 364, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hawken NM, Zaika EI, Nakagawa T, Engineering defined membrane-embedded elements of AMPA receptor induces opposing gating modulation by cornichon 3 and stargazin. J Physiol, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


