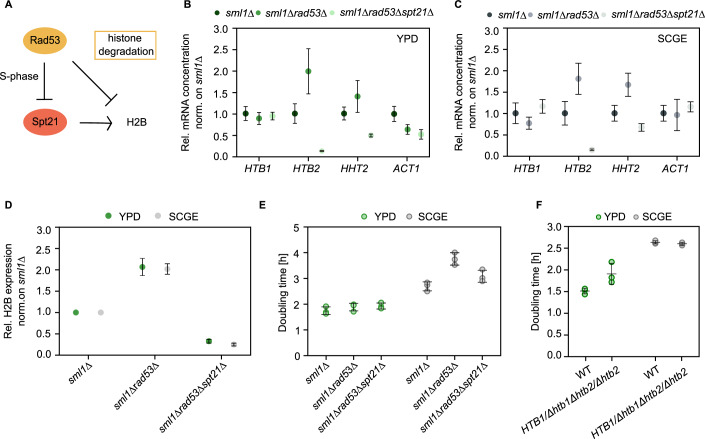
Figure 6. Cells growing on non-fermentable carbon sources are more sensitive to histone overexpression than in nutrient-rich conditions.
(A) Rad53 is required for the degradation of excess histones and also regulates histone levels by inhibiting the transcription activator Spt21. (B, C) mRNA concentrations of HTB1, HTB2, ACT1, and HHT2 (normalized on RDN18) for sml1Δrad53Δ and sml1Δrad53Δspt21Δ cells are shown as mean fold changes with respect to sml1Δ, in YPD (B) and SCGE (C). Error bars indicate standard errors across n = 3–5 biological replicates. (D) H2B protein levels, normalized on total protein, as determined by western blot analysis. Mean fold changes with respect to sml1Δ and standard errors of at least four replicates are shown. (E) Doubling times calculated from growth curves of sml1Δ, sml1Δrad53Δ, and sml1Δrad53Δspt21Δ cells growing on fermentable and non-fermentable carbon sources. Lines with error bars represent the mean and standard deviation of n = 3 independent measurements. (F) Reduced histone amounts slow down cell growth in rich nutrients. Doubling times were calculated from growth curves of HTB1/Δhtb1Δhtb2/Δhtb2 and wild-type diploid cells growing on fermentable and non-fermentable carbon sources. Lines and error bars represent the means and standard deviations of n = 3 independent measurements shown as individual dots. Source data are available online for this figure.