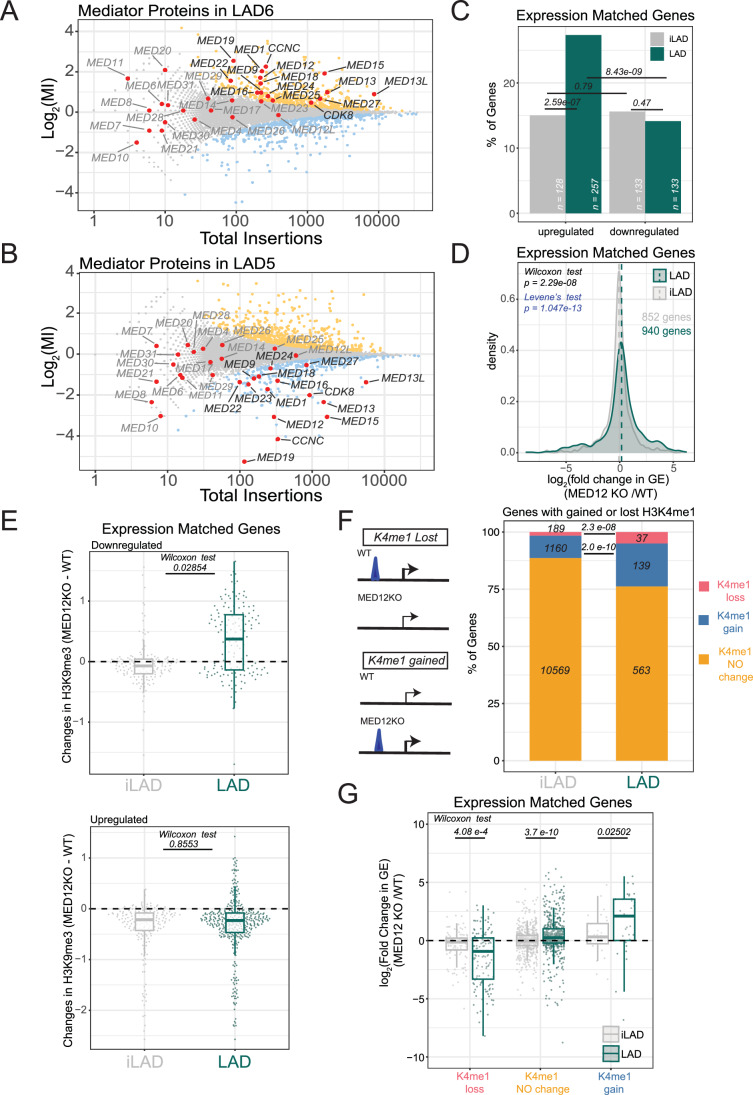
Figure 5. Mediator modulates heterochromatin and putative enhancers in LADs.
(A, B) Screen results with Mediator subunits highlighted in red. Mediator mainly acts as a repressor in LAD6 (A) and mostly as an activator in LAD5 (B). Blue, LAD activators; orange, LAD repressors; gray, not significant; red, Mediator subunits. (C) Percentage of up- and down-regulated genes following MED12 knockout for LAD and expression-matched iLAD genes. “n” indicates the number of genes analyzed in each class. Completely inactive genes (TPM < 0.3 in control sample) were not included in this analysis. Statistical significance was calculated with Fisher’s Exact test (D) log2(fold change) in gene expression (GE) following MED12 knockout for expression-matched LAD and iLAD genes. Statistical significance was calculated with Wilcoxon test for comparison of median, and Levene’s test for comparison of variance. (E) Changes in H3K9me3 levels at genes for downregulated (top) and upregulated (bottom) LAD genes or expression-matched iLAD genes. (F) Left: cartoon depicting how gain and loss of H3K4me1 peaks following MED12 knockout were selected. Right: Proportion of iLAD and LAD genes with a gain or loss of proximal H3K4me1 peak following MED12 knockout. P values according to Fisher’s Exact test. (G) log2(fold change) in gene expression (GE) following MED12 knockout for LAD and expression-matched iLAD genes, divided by gain or loss of proximal H3K4me1 peak following MED12 knockout. P values in (E, G) are according to Wilcoxon’s test. For (G, E), the central line in the boxplots represents the median. The lower and upper hinges correspond to the first and third quartiles (the 25th and 75th percentiles). The upper and lower whiskers extends from the hinge to the largest or smallest values respectively no further than 1.5 * inter-quartile range. Outliers were removed only for visualization purposes. For (B, C–F, G), data are from (Haarhuis et al, 2022) and results are from 6 biological replicates.