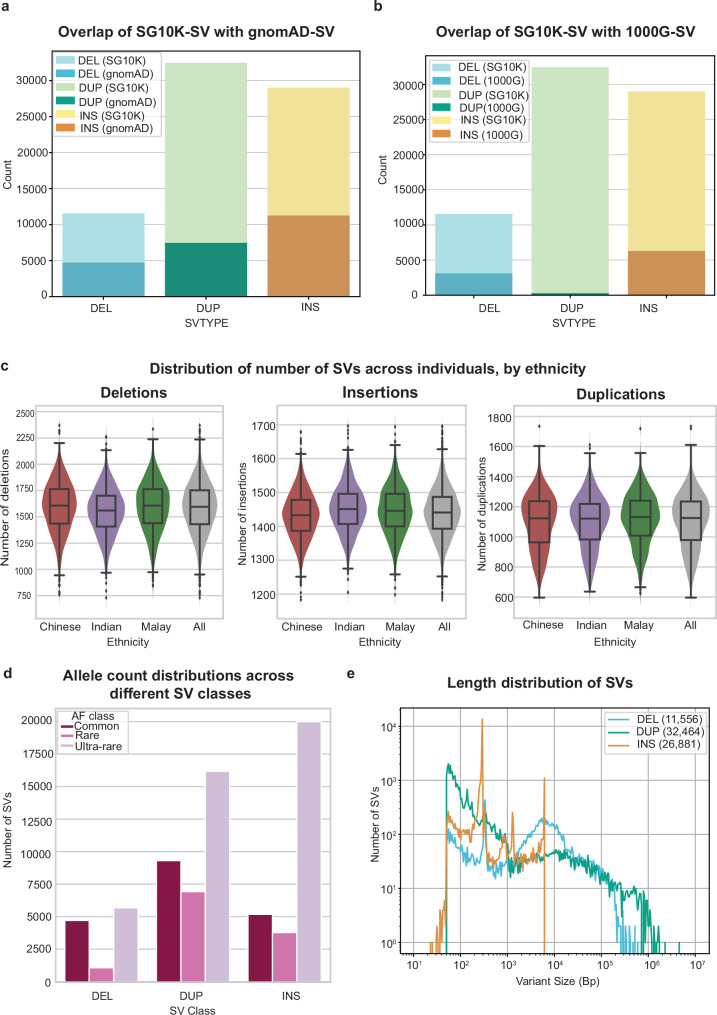
Fig. 2. SG10K-SV-r1.4 Structural Variant catalogue properties.
a Number of SG10K-SV-r1.4 variants detected in the discovery callset that overlap with gnomAD-SV. b Number of SG10K-SV-r1.4 variants detected in the discovery callset that overlap with 1000G-SV. c Violin plot and boxplot showing the number of SV per genome across individuals of different ethnicity group (3088 Chinese, 1237 Indians and 1144 Malays). The boxplot displays the minimum and maximum number of SVs as well as the median and the first/third quartile. DEL deletions, DUP duplications, INS insertions (including MEIs). d Number of SVs in different classes segregated by allele frequencies in the SG10K-SV-r1.4 discovery callset. The majority of the SVs are rare variants (AF < 1%). e Size distribution of SVs in SG10K-SV-r1.4 discovery callset. DEL deletions, DUP duplications, INS insertions (including MEIs). Expected Alu, SVA and LINE1 MEIs peaks at around 300 bp, 2100 bp and 6000 bp, respectively.