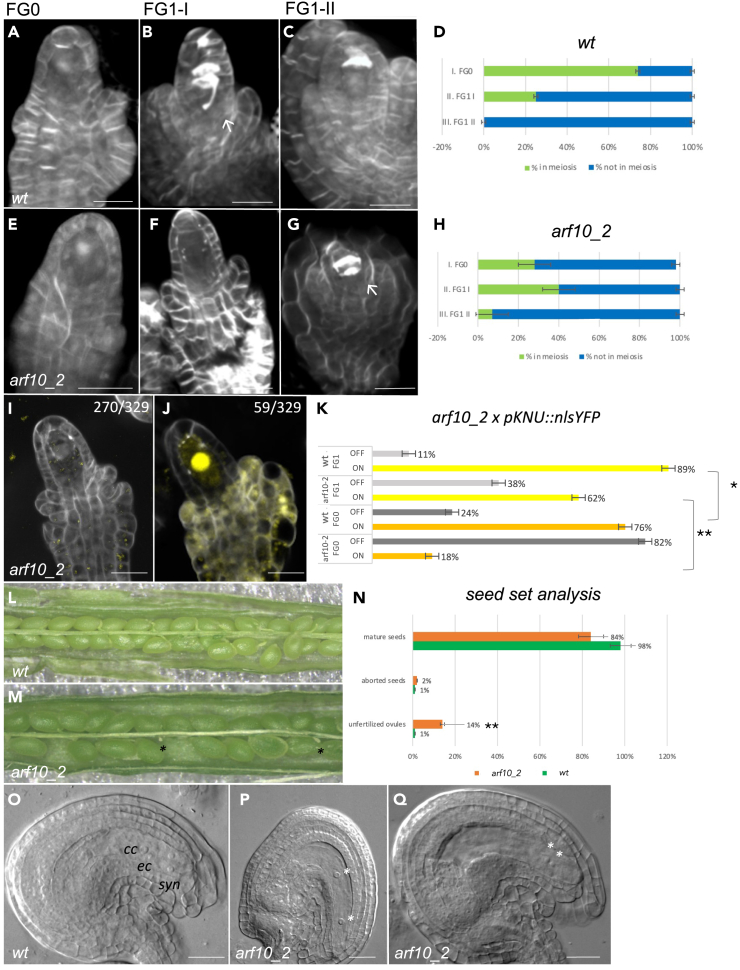
Figure 4.
arf10_2 defective mutant reproductive phenotype shows developmental delay
(A–C) wild type Renaissance staining highlighting meiotic progression at three different ovule stages; (A) FG0, finger-like ovule showing elongating integuments.
(B) FG1-I, meiosis ongoing or completed; the integuments have yet to cover the ovule.
(C) FG1-II after meiosis, integuments fully elongated, callose accumulation signifies tetrad degeneration.
(D) meiosis proportion in Renaissance-stained wild type ovules at three different stages.
(E–G) arf10-2 Renaissance staining highlighting meiotic progression at three different ovule stages (stages as in A–C). Meiosis is still in progress when integuments are fully elongated (white arrow).
(H) meiosis proportion in Renaissance-stained arf10_2 ovules at three different stages.
(I and J): arf10_2 ovules without (I) and with (J) pKNU:nlsYFP signal.
(K) Bar chart showing pKNU:nlsYFP expression in wild type and arf10-2 backgrounds at FG0 and FG1 stages: ON: YFP expression, OFF: no expression.
(L and M) seed set analysis in WT and arf10_2, black asterisks mark unfertilized ovules.
(N) proportion between mature seeds/aborted seeds/unfertilized ovules.
(O–Q) DIC imaging of wild type and arf10_2 mature ovules. Some arf10_2 ovules presented a block during gametogenesis; white asterisks indicate nuclei. The significance of differences between wild type and mutants for seed set and pKNU:YFP signal detection was evaluated by the Student’s t test (∗∗p < 0.05; ∗∗∗p < 0.001). For DIC imaging, the significance of statistical analyses was evaluated using the 95% Confidence Interval (CI): 15.15% vs. 2.33%; n = 396 and 300; 95% CI: 0.1184 < p < 0.1915 and 0.0102 < p < 0.0495 for arf10_2 and the wild type, respectively, Data are represented as mean ± SEM; scale bars: 20 μm; cc: central cell; ec: egg cell; syn: synergid cells.