Abstract
Fanconi anemia (FA) is an inherited disorder characterized by diverse congenital malformations, progressive pancytopenia, and predisposition to hematological malignancies and solid tumors. The role of the Fanconi anemia pathway in DNA repair mechanisms and genome instability is well studied. However, the consequences of inherited mutations in genes encoding the FA proteins and the acquired mutations due to impaired DNA repair complex in immune cells are far from understood. Patients with FA show bone marrow failure (BMF) and have a higher risk of developing myelodysplasia (MDS) or acute myeloid leukemia (AML) which are directly related to having chromosomal instability in hematopoietic stem cells and their subsequent progeny. However, immune dysregulation can also be seen in FA. As mature descendants of the common lymphoid progenitor line, NK cells taken from FA patients are dysfunctional in both NK cell-mediated cytotoxicity and cytokine production. The molecular bases for these defects are yet to be determined. However, recent studies have provided directions to define the cause and effect of inherited and acquired mutations in FA patients. Here, we summarize the recent studies in the hematopoietic dysfunction, focusing on the impairment in the development and functions of NK cells in FA patients, and discuss the possible mechanisms and future directions.
Keywords: NK cells, Fanconi anemia, DNA damage
I. INTRODUCTION
Pediatrician Guido Fanconi first described Fanconi anemia (FA) in 19271,2 when he treated a family in which 3 children had pancytopenia and congenital malformations. The disease manifested between the ages of 5 and 7, and all 3 died of a severe condition that resembled pernicious anemia.2 Detailed observations by Fanconi provided the basis for its clinical presentation and its earliest disease diagnosis. FA is a rare disease with an estimated 1 in 360,000 live births and a carrier frequency of approximately 1 in 181.3,4 FA can be challenging to diagnose due to a wide variety of symptoms presented by the patients.2 FA is characterized by diverse congenital and developmental defects, including short stature, microcephaly, abnormal skin pigmentation, and genitourinary and gastrointestinal abnormalities.5–9
At least 21 FA genes have been identified to date, and they are: FANCA, FANCB, FANCC, BRCA2/FANCD1, FANCD2, FANCE, FANCF, FANCG, FANCI, BRIP1/FANCJ, FANCL, FANCM, PALB2/FANCN, RAD51C/FANCO, SLX4/FANCP, ERCC4/FANCQ, RAD51/FANCR, BRCA1/FANCS, UBE2T/FANCT, XRCC2/FANCU, and MAD2L2/REV7/FANCV.10–13 FANCM, FANCO, FANCR, and FANCS are FA-like genes because they are not associated with bone marrow failure (BMF) in FA patients.14,15 FA has different complementation groups based on the mutated genes in FA patients.16,17 Approximately 85% of all patients in the International Fanconi Anemia Registry (IFAR) with a known complementation group are defective in one of the three most common disease-causing genes FANCA, FANCC, FANCG.18
Most FA groups are inherited in an autosomal recessive manner, but complementation Group B is X-linked recessive.19 FANCR gene dominant-negative mutation has been reported in patients with FA.20 Hematologic abnormalities occur in almost all FA patients, most presenting with BMF in the first decades of life.21 The BM microenvironment of patients with FA is hyperinflammed, related to increased levels of tumor necrosis factor-α (TNF-α) and interferon-γ (IFN-γ). This may have a role in the apoptosis of hematopoietic stem cells (HSCs) and, as a consequence, a significant impairment in lymphocyte development.15,22 In addition to adverse effects of IFN-γ, TNF-α, and other pro-inflammatory cytokines in FA patients, BMF may be exaggerated by the toxic effect of endogenous aldehyde and DNA damage-induced p53 activation, both of which result in HSC ablasion.15,23 One important outcome of this germline predisposition to DNA damage is the impaired ability of NK cell development and function.24–26
FA patients are monitored for myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML).27 Clonal evolution into MDS or AML occurs in 30% of FA patients by age 40, which frequently associates anomalies in a gain of 1q and 3q, deletion of 7q, and RUNX1 gene.28 The presence of aberration in chromosome 3 has been associated with hematopoietic malignancy. In these patients, the 3-year risk of MDS was 90%, and AML was 17%.15,29,30 FA patients also show a high predisposition to other types of cancers. The most common cancers in these patients are of the head and neck, breast, and gastrointestinal tract.11 This review will focus on the impairment in the hematopoietic compartment, emphasizing the developmental and functional defects of NK cells in FA patients.
II. FANCONI ANEMIA PATHWAY AND DNA REPAIR MECHANISM
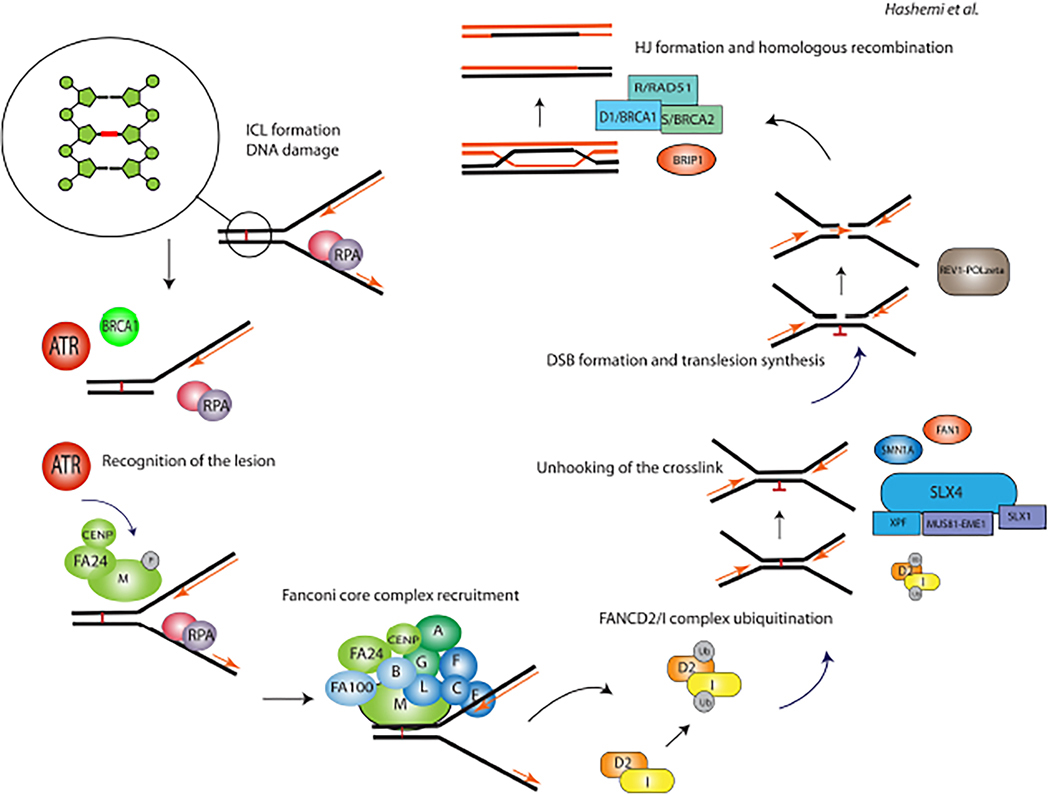
FA gene products are essential in repairing DNA interstrand crosslinks (ICLs) and regulating cellular responses to the genotoxic stress imposed by cell division. ICLs are covalent binding of 2 nucleotides in DNA strands caused by exogenous and endogenous factors that can impede DNA replication and transcription in cells.31 Due to these reasons, FA patients are susceptible to DNA damaging agents, and their DNA is sensitive to breakage when cultured with mitomycin C (MMC) or diepoxybutane (DEB), which results in G2/M arrest.32 The FA pathway is responsible for repairing ICLs and maintaining genome integrity. FANCM is a DNA-binding protein with helicase activity that recognizes the DNA lesions after phosphorylation by the ataxia-telangiectasia (ATM) and RAD3-related (ATR) checkpoint kinase.33 This protein senses the ICLs with the cooperation of other proteins, including FA-associated protein 24 (FAAP24) and the histone fold proteins MHF1 (also known as FAAP16 or CENPS) and MHF2 (also known as FAAP10 or CENPX) (Fig. 1).
FIG. 1:
Fanconi anemia pathway and DNA repair. ICLs are covalent binding of two nucleotides in DNA strands caused by exogenous and endogenous factors that can impede DNA replication and transcription in cells. The illustration depicts the pathway and the core complex associated with the Fanconi anemia repair pathway.
FANCM recruits the FA core complex, which is comprised of 14 proteins (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, FANCM, FANCT, FAAP100, MHF1, MHF2, FAAP20, and FAAP24).34 In some studies, the FANCM gene is excluded from the bonafide FA genes because loss-of-function FANCM variants are more common than initially predicted.13 FA core complex functions as a ubiquitin ligase for two other FA proteins, FANCD2 and FANCI, by the RING-finger containing E3 ubiquitin ligase, FANCL. Activation of FANCD2–FANCI complex triggers the DNA damage response. The FA pathway promotes ICLs repair through the incision of the lesion by nucleases such as FANCP (SLX4) and ERCC4, and the resulting DNA incision is repaired as BRCA2 promotes the formation of RAD-51-based nucleoprotein filaments on single-strand DNA for homologous recombination (Fig. 2). Deubiquitylation of the FANCD2-FANCI complex by USP1-UAF1 is required for the correct functioning of the FA pathway.35 Biallelic inactivation of any of the above key components of core complex results in FA.
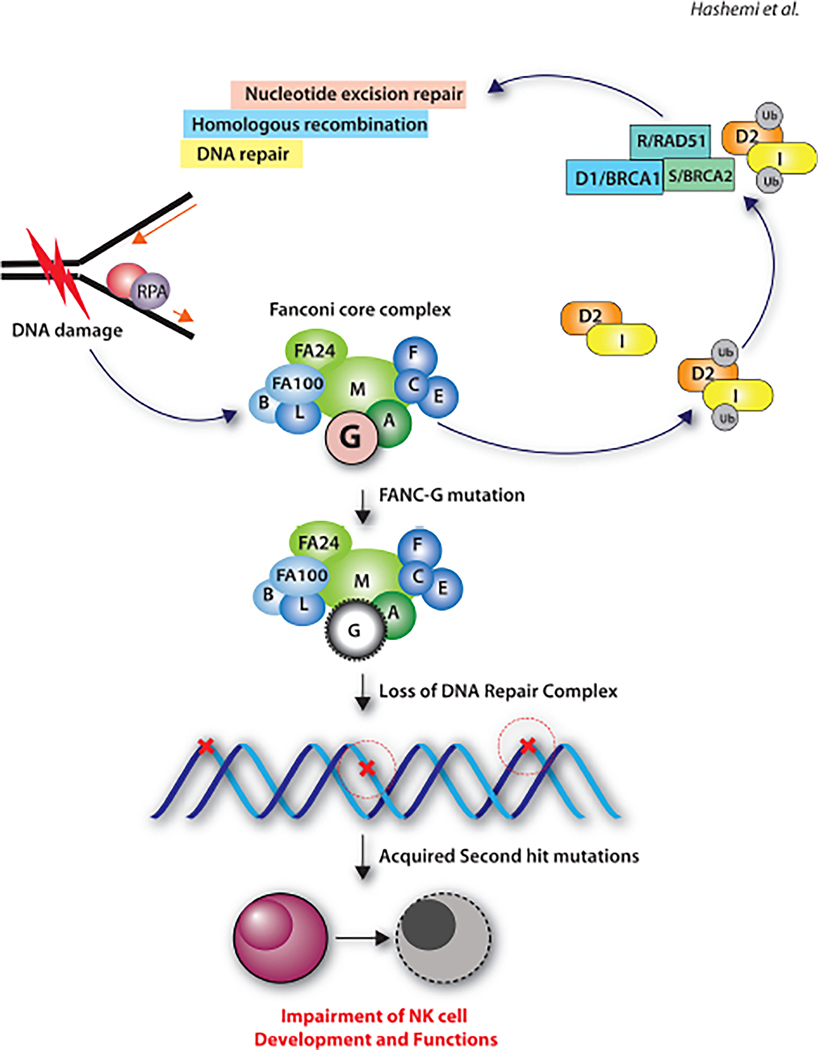
FIG. 2:
Potential mechanisms associated with impaired NK cell development and function in Fanconi anemia patients. Fanconi anemia subtype-G mutation as a model to illustrate the primary mutation in Fanconi anemia genes and the acquired second-hit mutations in NK cells. The second-hit mutations can lead to developmental and functional defects.
III. FANCONI ANEMIA AND IMMUNE DEFECTS
FA genes are involved in the physiological response to DNA damage. A failure of the FA complex results in BMF. Many children with FA invariably experience pancytopenia during the first decade of life, associated with stem cell loss in the hematopoietic compartment.36–40 BMF is the major cause of morbidity and mortality of FA.32,41 Patients with FA have a higher risk of developing MDS or AML. These indicate that mutations in FANC genes not only result in quantitative immune defects, but can also result in impaired immune cell development, differentiation, and functions.42
In order to better understand this DNA repair deficiency, murine models have been developed to analyze the impact of FA mutations on hematopoietic stem cells. However, these animal models have not faithfully replicated the clinical pathology seen in FA patients.43 FANC–/– mice did not present with any of the hematological impairments found in FA patients.44,45 Sequential, nonlethal doses of MMC caused a progressive decrease in all peripheral blood parameters of FANC–/– mice. These agents specifically target the BM compartment, with no effect on other proliferative tissues.45 Other studies indicate that FA knockout mouse models such as FANCA, FANCC, FANCD2, or FANCG do not display all the major FA clinical manifestations, such as spontaneous hematological defects and leukemia development.46 A potential reason for the disease discrepancies between FA patients and murine models could be the lack of inflammatory stress in the murine models.47 These complications with animal models possess a significant limitation in obtaining clinically-relevant knowledge. However, recent advances with humanized-mice models and advanced technologies, including single-cell transcriptomic profiling, provide opportunities to define the molecular basis of FA.
The immune system in FA patients can be defective in different ways. Increased apoptosis could be due to a higher level of the death receptor Fas (CD95) in CD34+ stem cells from patients with FA.47 Conversely, overexpression of the FANCC protein suppresses apoptosis in CD34+ stem cells from FANCC patients.48 Several studies have investigated lymphocyte function and development in FA patients.24,49–52 Myers et al. reported quantitative and functional evaluation of immune function in 10 children with FA. Total numbers of B and NK cells were decreased compared to controls, while the T cell numbers were within the expected range in these patients.53 Another study from the same group indicates a lower number of CD4+ T cells in a larger FA cohort who have not undergone bone marrow transplantation. In this study, they assessed immune cell numbers and function in 29 FA patients. B cells, memory B cells, and the serum levels of IgM were decreased in these patients, while CD8+ T cell and absolute T cell numbers were comparable to that of healthy controls. Cytotoxic T cells and antigen-specific proliferation response to tetanus and candida were also diminished in these patients.54 However, there has not been consensus across studies; for example, another study states that CD8+ T cells were lower in FA patients compared to healthy controls, leading to an increased ratio of CD4:CD8 T cells.55
In addition to lymphocytes, monocytes and macrophages also show impaired functions in the murine models of FA.56 Liu et al. examined FANCC−/− mice for macrophage functions. Their study indicates impaired cytoskeletal rearrangements in FANCC−/− macrophages and altered monocyte/macrophage trafficking in vivo. FANCC−/− peritoneal macrophages displayed impaired adhesion to fibronectin or endothelial cells, reduced chemoattractant-mediated migration, and decreased phagocytosis. Moreover, dysregulated F-actin rearrangement was detected in FANCC−/− macrophages after adhesion to fibronectin.56 FA patients show dysregulation of immune factors like cytokines and chemokines. The elevated level of TNF-α in FA patients led to the accumulation of inflammatory reactive oxygen species (ROS) and deregulated stress kinase (p38 and JNK) activation in FA bone marrow cells.42 ROS accumulation and kinase activation initiate apoptosis of bone marrow cells and clonal proliferation, leading to BMF, MDS, or AML in FA patients. The increased secretion of TNF-α and altered production of other growth factors and cytokines, including reduced expression of interleukin-6 (IL-6) and granulocyte-macrophage colony-stimulating factor (GM-CSF), may change the BM microenvironment by leading to factor deprivation or constant exposure to mitogenic inhibitors.57 These studies indicate that the dysfunction of immune cells in FA patients; however, the molecular basis of this immune dysfunction is yet to be fully understood.
IV. IMPAIRED NK CELLS DEVELOPMENT AND FUNCTION IN FANCONI ANEMIA
FA patients have been reported to have reduced absolute numbers and the percentages of NK cells.25,26,37 Immunological assessment in a large FA cohort of 49 patients who have not undergone hematopoietic stem cell transplantation or developed malignancies showed the number of NK cells reduced significantly compared to healthy individuals.54 However, in another study evaluating 42 FA patients, there was no difference noted in the total number of NK cells, while the proportions of CD56bright and CD56dim NK subsets were changed.55 This study found an increase in the CD56bright NK subset in FA patients compared to healthy controls. Additionally, CD56dim NK cells showed decreased CD16 expression, which is an important marker for cytotoxicity. Reduction in NK cell numbers and decreased cytotoxic potentials have a causal relationship with increased susceptibility to infections, including flaviviruses, viral hepatitis, influenza, and HIV.58 Thus, acute or chronic viral infections seen in FA patients could be due to not only a reduced NK cell number but also their effector functions.
NK cells mediate their cytotoxic functions against transformed or infected cells, primarily utilizing the perforin/granzyme pathway.59 Recent studies have described a defective degranulation mechanism as the cause of impaired NK-cell cytotoxicity in FA patients.25,26,37 Flow cytometry-based analyses found that while the intracellular perforin was of expected levels, the degranulation of cytotoxic vesicles based on the cell surface expression of CD107a was significantly reduced.26 These findings indicate that a defect in the DNA repair complex could have resulted in acquired mutations in one of the essential proteins involved in the sorting and trafficking of the cytotoxic granule-containing vesicles or the Fanconi proteins themselves involved in this exocytosis pathway. Recent studies have shown that FA proteins are involved in cellular functions apart from non-DNA repair, including immune response.60–62 The residual NK cell activity in FA patients could ironically also harm their hematopoietic cells while protecting them from chronic infections. NKG2D provides sufficient stimulus to activate cytolysis and cytokine production by interacting with their ligands, including MIC-A, MIC-B, and ULBPs.63,64 Earlier work showed that mouse and human NKG2D ligands are upregulated in non-tumor cell lines by exposure to genotoxic stress and drugs that stall DNA replication, such as MMC, hydroxyurea, and 5-fluorouracil.65 Following exposure to these pharmacological agents, major DNA damage checkpoint pathways, initiated by mutated ATM and ATR (ATM- and Rad3-related) protein kinases are activated.65 This ligand upregulation within the hematopoietic compartment can make these cells targets of NK cell-mediated cytotoxicity. Thus, genotoxicity resulting from DNA damage can mount an immune response from NK cells leading to the loss of hematopoietic cells. A cautious analysis is required to determine whether autologous NK cells commit this fraternal killing.
How an impairment in the DNA repair pathway leads to developmental and functional defects in NK cells in FA patients is not understood. Controlled DNA breakage is an essential mechanism for normal differentiation and maturation of T and B cells during their ontogeny. A defect in this process at an early age in individuals could result in impaired adaptive immunity with pathological consequences. In contrast to cytotoxic T cells, NK cells neither utilize clonotypic receptors nor require prior antigen exposure to mediate their anti-tumor effects.66,67 RAG1 and RAG2 genes are involved in immunoglobulin V-D-J in B cell receptor and T cell receptor recombinations. Defects in RAG1 and RAD2 genes cause several diseases.68 A recent study indicated that RAG1 and RAG2 might play essential roles in NK cells.66,69 NK cells that lacked RAG expression have a decreased expression in essential components of the DNA damage response, including DNA-PKcs (PRKDC), Ku80 (XRCC5), Chk2 (CHEK2), and ATM (ATM) and inefficient DNA repair in mature cells. Similar issues in repairing DNA damage and an increased inflammatory response were found in the inherited disease Ataxia-Telangiectasia syndrome, a cancer predisposition syndrome.70 Thus, using these examples, impairment in the DNA damage pathway could have an impact on the development and functionality of NK cells.
Acquired mutations can occur in transcription factors or functional genes of NK cells in FA patients. Defects in transcription factors can lead to progressive loss of NK progenitors and thereby a reduced number of mature NK cells. If genes that encode functional proteins such as perforin and granzymes acquire mutations, this could lead to impaired effector functions. Deleterious mutations in genes encoding perforin or granzyme-b can result in impaired NK cell-mediated cytotoxicity. However, earlier studies have not reported such acquired mutations due to the defective DNA repair complex in FA patients. The proposed non-canonical functions of FA proteins hold the promise to explain the wide range of heterogeneous clinical manifestations seen in these patients. Therefore, novel cellular functions ascribed to FA proteins may provide mechanistic explanations.
Earlier studies have shown that the core DNA complex comprised of FANCA, FANCC, FANCD2, FANCF, and FANCG may interact with non-FA proteins such as the Sorting Nexin protein (SNX5), Brahma-Related Gene 1 protein (BRG1), IκB Kinase-2 (IKK2), Breast Cancer 1 (BRCA1) and is involved in various cellular functions.71–74 Murine FANCD2 localizes within mitochondrion and associates with the nucleoid complex components ATAD3 and TUFM.74 Interaction between FANCA and SNX5 may provide a direct molecular mechanism for defective vesicle trafficking, thereby impairing NK cell-mediated cytotoxicity.75 Heterogeneous clinical pathology makes it challenging to identify a common molecular basis for the defects seen in all FA patients. However, detailed analyses at the transcriptome level may provide pertinent molecular mechanisms in a subset of FA patients.
NK cells also generate significant quantities of pro-inflammatory cytokines and chemokines.59,76 NK cells produce inflammatory cytokines in response to activation receptor stimulation (NKG2D, NCR1, NCR2, NCR3, CD244, and KIRs) and cytokine-induced activation (IL-12, IL-18, IL-23, IL-27, and IL-35) signaling.77,78 In contrast to a defect in the cellular cytotoxicity, FA patients present an inflammatory pathology.71,72 Earlier studies describe a significant increase in IFN-γ, IL-6, IL-10, IL-7 GM-CSF, MCP-1, MIP-1α, MIP-1β, and IP10/CXCL10 levels in FA patients compared to healthy controls, which provides the basis for an augmented inflammatory phenotype.26 The hyperinflammatory pathology in FA patients could be due to the presence of excess nucleic acid loads in cellular compartments that permit recognition by proteins designed to recognize pathogen-associated molecular patterns, including toll-like receptors (TLR).79 Indeed, loss of the FANCP or FANCD2 genes resulted in the accumulation of cytoplasmic DNA, leading to the stimulation of the cGAS-STING pathway and type-1 interferon production.79 Human and murine NK cells express several TLRs.80–85 Thus, initiation of NK cell activation via either cell surface or intracellular innate DNA sensors could potentially result in the hyperproduction of pro-inflammatory cytokines. Detailed molecular analyses are needed to precisely identify the signaling pathways and transcriptional regulation of this immune pathology in FA patients.
V. SUMMARY AND FUTURE DIRECTIONS
DNA damage initiates cell cycle arrest, DNA repair, and apoptosis, but its role in immune functionality is unknown. FA is an inherited disease with a deficiency in DNA damage response leading to BMF. FA patients undergo progressive BMF during childhood, which requires allogeneic hematopoietic stem cell transplantation. The genome instability in FA patients results in the accumulation of newer mutations that affect the maintenance and differentiation of hematopoietic stem cells and their progeny. The total number, subset ratio, and functional capabilities of NK cells are significantly reduced in FA patients compared to healthy individuals. The cytotoxic potentials of NK cells are diminished in FA patients, while their inflammatory responses are augmented. While the role of FA gene products for the regulation of DNA repair mechanisms has been well-established, there remains little insight to explain how NK cell impairment is linked to this defect. Does an impairment in the DNA repair pathway initiate the NK cell impairment in these patients? Or has the acquisition of secondary mutations led this to immune cell dysfunction, including defects seen in NK cells? Do the FANC proteins mediate non-DNA repair-related functions essential for NK cell development, survival, and effector functions? Answers to these essential questions will provide a mechanistic explanation for NK cell impairment and the overall immune dysfunctions in FA patients.
ACKNOWLEDGMENTS
We dedicate this work to our inspiring colleague Dr. Mathew Riese, MD, PhD, who passed away young in December 2020. This work was supported in part by NIH R01 AI102893 and NCI R01 CA179363 (S.M.); HRHM Program of MACC Fund (S.M.), Nicholas Family Foundation (S.M.); Gardetto Family (S.M.); MCW-Cancer Center-Large Seed Grant (S.M. and M.S.T.); MACC Fund (S.M.); Ann’s Hope Melanoma Foundation (S.M.); and Advancing Healthier Wisconsin (S.M.).
ABBREVIATIONS:
- AML
acute myeloid leukemia
- BMF
bone marrow failure
- FA
Fanconi anemia
- HSC
hematopoietic stem cells
- ICLs
interstrand crosslinks
- IFAR
International Fanconi Anemia Registry
- IFN-γ
interferon-gamma
- IL
interleukin
- MDS
myeloid dysplasia
- NK
natural killer cells
REFERENCES
- 1.Lobitz S, Velleuer E. Guido Fanconi (1892–1979): A jack of all trades. Nature Rev Cancer. 2006;6(11):893–8. [DOI] [PubMed] [Google Scholar]
- 2.Mamrak NE, Shimamura A, Howlett NG. Recent discoveries in the molecular pathogenesis of the inherited bone marrow failure syndrome Fanconi anemia. Blood Rev. 2017;31(3):93–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenberg PS, Tamary H, Alter BP. How high are carrier frequencies of rare recessive syndromes? Contemporary estimates for Fanconi Anemia in the United States and Israel. Am J Med Genet A. 2011. Aug;155A(8):1877–83. PubMed PMID: 21739583. PMCID: PMC3140593. Epub 2011/07/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinberg-Shemer O, Goldberg TA, Yacobovich J, Levin C, Koren A, Revel-Vilk S, Ben-Ami T, Kuperman AA, Shkalim Zemer V, Toren A, Kapelushnik J, Ben-Barak A, Miskin H, Krasnov T, Noy-Lotan S, Dgany O, Tamary H. Characterization and genotype-phenotype correlation of patients with Fanconi anemia in a multi-ethnic population. Haematologica. 2019. Sep 26. PubMed PMID: 31558676. Epub 2019/09/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shahidi NT. Fanconi anemia, dyskeratosis congenita, and WT syndrome. Am J Med Genet Suppl. 1987;3:263–78. PubMed PMID: 2453204. Epub 1987/01/01. [DOI] [PubMed] [Google Scholar]
- 6.Chaganti RS, Houldsworth J. Fanconi anemia: A pleotropic mutation with multiple cellular and developmental abnormalities. Ann Genet. 1991;34(3–4):206–11. PubMed PMID: 1809228. Epub 1991/01/01. [PubMed] [Google Scholar]
- 7.Mehta PA, Tolar J. Fanconi Anemia. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, editors. GeneReviews® [Internet]. Seattle, WA: University of Washington; 1993. [PubMed] [Google Scholar]
- 8.dos Santos CC, Gavish H, Buchwald M. Fanconi anemia revisited: Old ideas and new advances. Stem Cells. 1994. Mar;12(2):142–53. PubMed PMID: 8199559. Epub 1994/03/01. [DOI] [PubMed] [Google Scholar]
- 9.Auerbach AD. Fanconi anemia. Dermatol Clin. 1995. Jan;13(1):41–9. PubMed PMID: 7712649. Epub 1995/01/01. [PubMed] [Google Scholar]
- 10.Nepal M, Che R, Ma C, Zhang J, Fei P. FANCD2 and DNA damage. Int J Mol Sci. 2017;18(8):1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nepal M, Che R, Zhang J, Ma C, Fei P. Fanconi anemia signaling and cancer. Trends Cancer. 2017;3(12):840–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouffard F, Plourde K, Bélanger S, Ouellette G, Labrie Y, Durocher F. Analysis of a FANCE splice isoform in regard to DNA repair. J Mol Biol. 2015;427(19):3056–73. [DOI] [PubMed] [Google Scholar]
- 13.Bogliolo M, Surrallés J. Fanconi anemia: A model disease for studies on human genetics and advanced therapeutics. Curr Op Genet Develop. 2015;33:32–40. [DOI] [PubMed] [Google Scholar]
- 14.Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, Neveling K, Endt D, Kesterton I, Autore F. Mutation of the RAD51C gene in a Fanconi anemia–like disorder. Nat Genet. 2010;42(5):406–9. [DOI] [PubMed] [Google Scholar]
- 15.Savage SA, Walsh MF. Myelodysplastic syndrome, acute myeloid leukemia, and cancer surveillance in Fanconi anemia. Hematol Oncol Clin North Am. 2018;32(4):657–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Che R, Zhang J, Nepal M, Han B, Fei P. Multifaceted Fanconi anemia signaling. Trends Genet. 2018;34(3):171–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross) linked to DNA repair. Cell. 2005;123(7):1191–8. [DOI] [PubMed] [Google Scholar]
- 18.Auerbach AD. Fanconi anemia and its diagnosis. Mutat Res. 2009;668(1–2):4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meetei AR, Levitus M, Xue Y, Medhurst AL, Zwaan M, Ling C, Rooimans MA, Bier P, Hoatlin M, Pals G. X-linked inheritance of Fanconi anemia complementation group B. Nat Genet. 2004;36(11):1219–24. [DOI] [PubMed] [Google Scholar]
- 20.Ameziane N, May P, Haitjema A, Van De Vrugt HJ, van Rossum-Fikkert SE, Ristic D, Williams GJ, Balk J, Rockx D, Li H. A novel Fanconi anaemia subtype associated with a dominant-negative mutation in RAD51. Nat Commun. 2015;6(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore CA, Krishnan K. Bone Marrow Failure. [Updated 2020 Jul 13]. In: StatPearls [Internet]. Treasure Island, FL: StatPearls Publishing; 2021. Available from: https://www.ncbi.nlm.nih.gov/books/NBK459249/. [Google Scholar]
- 22.Young NS, Calado RT, Scheinberg P. Current concepts in the pathophysiology and treatment of aplastic anemia. Blood. 2006;108(8):2509–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ceccaldi R, Parmar K, Mouly E, Delord M, Kim JM, Regairaz M, Pla M, Vasquez N, Zhang Q-S, Pondarre C. Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 2012;11(1):36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fagerlie SR, Bagby GC. Immune defects in Fanconi anemia. Crit Rev Immunol. 2006;26(1):81–96. PubMed PMID: 16472069. Epub 2006/02/14. [DOI] [PubMed] [Google Scholar]
- 25.Myers KC, Sauter S, Zhang X, Bleesing JJ, Davies SM, Wells SI, Mehta PA, Kumar A, Marmer D, Marsh R, Brown D, Butsch Kovacic M. Impaired immune function in children and adults with Fanconi anemia. Pediatr Blood Cancer. 2017. Nov;64(11). PubMed PMID: 28557197. PMCID: PMC5639938. Epub 2017/05/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shabrish S, Kelkar M, Chavan N, Desai M, Bargir U, Gupta M, Mehta P, Chichra A, Chandrakala S, Taur P, Saxena V, Vundinti BR, Madkaikar M. Natural killer cell degranulation defect: A cause for impaired NK-cell cytotoxicity and hyperinflammation in fanconi anemia patients. Front Immunol. 2019;10:490. PubMed PMID: 30949167. PMCID: PMC6438155. Epub 2019/04/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peffault de Latour R, Soulier J. How I treat MDS and AML in Fanconi anemia. Blood. 2016;127(24):2971–9. [DOI] [PubMed] [Google Scholar]
- 28.Debureaux P, de Fontbrune FS, Bonfim C, Dalle J, Buchbinder N, Bertrand Y, Renard C, Forcade E, Fernandes J, Talbot A. FLAG-sequential regimen followed by bone marrow transplantation for myelodysplastic syndrome or acute leukemia in patients with Fanconi anemia: A Franco-Brazilian study. Bone Marrow Transplant. 2021;56(1):285–8. [DOI] [PubMed] [Google Scholar]
- 29.Tönnies H, Huber S, Kü Jr-S, Gerlach A, Ebell W, Neitzel H. Clonal chromosomal aberrations in bone marrow cells of Fanconi anemia patients: Gains of the chromosomal segment 3q26q29 as an adverse risk factor. Blood. 2003;101(10):3872–4. [DOI] [PubMed] [Google Scholar]
- 30.Cioc AM, Wagner JE, MacMillan ML, DeFor T, Hirsch B. Diagnosis of myelodysplastic syndrome among a cohort of 119 patients with fanconi anemia: Morphologic and cytogenetic characteristics. Am J Clin Pathol. 2010;133(1):92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y, Li L. DNA crosslinking damage and cancer-a tale of friend and foe. Translational Cancer Res. 2013;2(3):144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bagby GC. The genetic basis of fanconi anemia. In: Molecular mechanisms of fanconi anemia. Springer; 2006. p. 13–27. [Google Scholar]
- 33.Xue X, Sung P, Zhao X. Functions and regulation of the multitasking FANCM family of DNA motor proteins. Genes Dev. 2015;29(17):1777–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niraj J, Färkkilä A, D’Andrea AD. The Fanconi anemia pathway in cancer. Annual Rev Cancer Biol. 2019;3:457–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodríguez A, D’Andrea A. Fanconi anemia pathway. Curr Biol. 2017;27(18):R986–R8. [DOI] [PubMed] [Google Scholar]
- 36.Dalle JH. HSCT for Fanconi anemia in children: Factors that influence early and late results. Bone Marrow Transplant. 2008. Oct;42(Suppl 2):S51–3. PubMed PMID: 18978745. Epub 2008/11/26. [DOI] [PubMed] [Google Scholar]
- 37.Korthof ET, Svahn J, Peffault de Latour R, Terranova P, Moins-Teisserenc H, Socie G, Soulier J, Kok M, Bredius RG, van Tol M, Jol-van der Zijde EC, Pistorio A, Corsolini F, Parodi A, Battaglia F, Pistoia V, Dufour C, Cappelli E. Immunological profile of Fanconi anemia: A multicentric retrospective analysis of 61 patients. Am J Hematol. 2013. Jun;88(6):472–6. PubMed PMID: 23483621. Epub 2013/03/14. [DOI] [PubMed] [Google Scholar]
- 38.Risitano AM, Marotta S, Calzone R, Grimaldi F, Zatterale A, Contributors R. Twenty years of the Italian Fanconi anemia registry: Where we stand and what remains to be learned. Haematologica. 2016. Mar;101(3):319–27. PubMed PMID: 26635036. PMCID: PMC4815723. Epub 2015/12/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ayas M. Hematopoietic cell transplantation in Fanconi anemia and dyskeratosis congenita: A minireview. Hematol Oncol Stem Cell Ther. 2017. Dec;10(4):285–9. PubMed PMID: 28644950. Epub 2017/06/24. [DOI] [PubMed] [Google Scholar]
- 40.Degan P, Cappelli E, Regis S, Ravera S. New insights and perspectives in Fanconi anemia research. Trends Mol Med. 2019. Mar;25(3):167–70. PubMed PMID: 30744929. Epub 2019/02/13. [DOI] [PubMed] [Google Scholar]
- 41.Fagerlie SR, Diaz J, Christianson TA, McCartan K, Keeble W, Faulkner GR, Bagby GC. Functional correction of FA-C cells with FANCC suppresses the expression of interferon γ–inducible genes. Blood. 2001;97(10):3017–24. [DOI] [PubMed] [Google Scholar]
- 42.Du W, Erden O, Pang Q. TNF-α signaling in Fanconi anemia. Blood Cells Mol Dis. 2014;52(1):2–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parmar K, D’Andrea A, Niedernhofer LJ. Mouse models of Fanconi anemia. Mutat Res. 2009. Jul 31;668(1–2):133–40. PubMed PMID: 19427003. PMCID: PMC2778466. Epub 2009/05/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen M, Tomkins DJ, Auerbach W, McKerlie C, Youssoufian H, Liu L, Gan O, Carreau M, Auerbach A, Groves T, Guidos CJ, Freedman MH, Cross J, Percy DH, Dick JE, Joyner AL, Buchwald M. Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anaemia. Nat Genet. 1996. Apr;12(4):448–51. PubMed PMID: 8630504. Epub 1996/04/01.eng. [DOI] [PubMed] [Google Scholar]
- 45.Carreau M, Gan OI, Liu L, Doedens M, McKerlie C, Dick JE, Buchwald M. Bone marrow failure in the Fanconi anemia group C mouse model after DNA damage. Blood. 1998;91(8):2737–44. [PubMed] [Google Scholar]
- 46.Walter D, Lier A, Geiselhart A, Thalheimer FB, Huntscha S, Sobotta MC, Moehrle B, Brocks D, Bayindir I, Kaschutnig P, Muedder K, Klein C, Jauch A, Schroeder T, Geiger H, Dick TP, Holland-Letz T, Schmezer P, Lane SW, Rieger MA, Essers MA, Williams DA, Trumpp A, Milsom MD. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015. Apr 23;520(7548):549–52. PubMed PMID: 25707806. Epub 2015/02/25. [DOI] [PubMed] [Google Scholar]
- 47.Otsuki T, Nagakura S, Wang J, Bloom M, Grompe M, Liu JM. Tumor necrosis factor-α and CD95 ligation suppress erythropoiesis in Fanconi anemia C gene knockout mice. J Cellular Physiol. 1999;179(1):79–86. [DOI] [PubMed] [Google Scholar]
- 48.Walsh CE, Nienhuis AW, Samulski RJ, Brown MG, Miller JL, Young NS, Liu JM. Phenotypic correction of Fanconi anemia in human hematopoietic cells with a recombinant adeno-associated virus vector. J Clin Invest. 1994;94(4):1440–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamashita T, Nakahata T. Current knowledge on the pathophysiology of Fanconi anemia: From genes to phenotypes. Int J Hematol. 2001. Jul;74(1):33–41. PubMed PMID: 11530803. Epub 2001/09/04. [DOI] [PubMed] [Google Scholar]
- 50.Velez-Ruelas MA, Martinez-Jaramillo G, Arana-Trejo RM, Mayani H. Hematopoietic changes during progression from Fanconi anemia into acute myeloid leukemia: Case report and brief review of the literature. Hematology. 2006. Oct;11(5):331–4. PubMed PMID: 17607582. Epub 2007/07/04. [DOI] [PubMed] [Google Scholar]
- 51.Muller LU, Williams DA. Finding the needle in the hay stack: Hematopoietic stem cells in Fanconi anemia. Mutat Res. 2009. Jul 31;668(1–2):141–9. PubMed PMID: 19508850. PMCID: PMC2815349. Epub 2009/06/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith AR, Wagner JE. Current clinical management of Fanconi anemia. Expert Rev Hematol. 2012. Oct;5(5):513–22. PubMed PMID: 23146055. Epub 2012/11/14. [DOI] [PubMed] [Google Scholar]
- 53.Myers KC, Bleesing JJ, Davies SM, Zhang X, Martin LJ, Mueller R, Harris RE, Filipovich AH, Kovacic MB, Wells SI. Impaired immune function in children with Fanconi anaemia. British J Haematol. 2011;154(2):234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Myers KC, Sauter S, Zhang X, Bleesing JJ, Davies SM, Wells SI, Mehta PA, Kumar A, Marmer D, Marsh R. Impaired immune function in children and adults with Fanconi anemia. Pediatr Blood Cancer. 2017;64(11):e26599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Justo GA, Bitencourt MA, Pasquini R, Castelo-Branco MT, Almeida-Oliveira A, Diamond HR, Rumjanek VM. Immune status of Fanconi anemia patients: Decrease in T CD8 and CD56 dim CD16+ NK lymphocytes. Annals Hematol. 2014;93(5):761–7. [DOI] [PubMed] [Google Scholar]
- 56.Liu Y, Ballman K, Li D, Khan S, Derr-Yellin E, Shou W, Haneline LS. Impaired function of Fanconi anemia type C-deficient macrophages. J Leukocyte Biol. 2012;91(2):333–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du W, Adam Z, Rani R, Zhang X, Pang Q. Oxidative stress in Fanconi anemia hematopoiesis and disease progression. Antioxid. Redox Signal. 2008;10(11):1909–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mandal A, Viswanathan C. Natural killer cells: In health and disease. Hematol Oncol Stem Cell Ther. 2015;8(2):47–55. [DOI] [PubMed] [Google Scholar]
- 59.Abel AM, Yang C, Thakar MS, Malarkannan S. Natural killer cells: Development, maturation, and clinical utilization. Front Immunol. 2018;9:1869. PubMed PMID: 30150991. PMCID: PMC6099181. Epub 2018/08/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kupfer GM, Naf D, D’Andrea AD. Molecular biology of Fanconi anemia. Hematol Oncol Clin North Am. 1997. Dec;11(6):1045–60. PubMed PMID: 9443045. Epub 1998/01/27. [DOI] [PubMed] [Google Scholar]
- 61.Taniguchi T, D’Andrea AD. Molecular pathogenesis of Fanconi anemia: Recent progress. Blood. 2006. Jun 1;107(11):4223–33. PubMed PMID: 16493006. Epub 2006/02/24. [DOI] [PubMed] [Google Scholar]
- 62.Kee Y, D’Andrea AD. Molecular pathogenesis and clinical management of Fanconi anemia. J Clin Invest. 2012. Nov;122(11):3799–806. PubMed PMID: 23114602. PMCID: PMC3484428. Epub 2012/11/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Molfetta R, Quatrini L, Santoni A, Paolini R. Regulation of NKG2D-dependent NK cell functions: The yin and the yang of receptor endocytosis. Int J Mol Sci. 2017;18(8):1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jelenčić V, Šestan M, Kavazović I, Lenartić M, Marinović S, Holmes TD, Prchal-Murphy M, Lisnić B, Sexl V, Bryceson YT. NK cell receptor NKG2D sets activation threshold for the NCR1 receptor early in NK cell development. Nature Immunol. 2018;19(10):1083–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436(7054):1186–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hilton HG, Parham P. Missing or altered self: Human NK cell receptors that recognize HLA-C. Immunogenetics. 2017;69(8–9):567–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Konjević G, Vuletić A, Martinović KM. Natural killer cell receptors: Alterations and therapeutic targeting in malignancies. Immunol.Res. 2016;64(1):25–35. [DOI] [PubMed] [Google Scholar]
- 68.Karo JM, Schatz DG, Sun JC. The RAG recombinase dictates functional heterogeneity and cellular fitness in natural killer cells. Cell. 2014;159(1):94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coscoy L, Raulet DH. DNA mismanagement leads to immune system oversight. Cell. 2007;131(5):836–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Härtlova A, Erttmann SF, Raffi FA, Schmalz AM, Resch U, Anugula S, Lienenklaus S, Nilsson LM, Kröger A, Nilsson JA, Ek T, Weiss S, Gekara NO. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 2015. Feb 17;42(2):332–43. PubMed PMID: 25692705. Epub 2015/02/19.eng. [DOI] [PubMed] [Google Scholar]
- 71.Cheung RS, Taniguchi T. Recent insights into the molecular basis of Fanconi anemia: Genes, modifiers, and drivers. Int J Hematol. 2017. Sep;106(3):335–44. PubMed PMID: 28631178. PMCID: PMC5904331. Epub 2017/06/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bagby G. Recent advances in understanding hematopoiesis in Fanconi anemia. F1000Res. 2018;7:105. PubMed PMID: 29399332. PMCID: PMC5785713. Epub 2018/02/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen X, Bosques L, Sung P, Kupfer GM. A novel role for non-ubiquitinated FANCD2 in response to hydroxyurea-induced DNA damage. Oncogene. 2016. Jan 7;35(1):22–34. PubMed PMID: 25893307. PMCID: PMC5450631. Epub 2015/04/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang T, Du W, Wilson AF, Namekawa SH, Andreassen PR, Meetei AR, Pang Q. FANCD2 in vivo interaction network reveals a non-canonical role in mitochondrial function. Sci Rep. 2017. Apr 5;7:45626. PubMed PMID: 28378742. PMCID: PMC5381226. Epub 2017/04/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Otsuki T, Kajigaya S, Ozawa K, Liu JM. SNX5, a new member of the sorting nexin family, binds to the Fanconi anemia complementation group A protein. Biochem Biophys Res Commun. 1999. Nov 30;265(3):630–5. PubMed PMID: 10600472. Epub 1999/12/22. [DOI] [PubMed] [Google Scholar]
- 76.Rajasekaran K, Kumar P, Schuldt KM, Peterson EJ, Vanhaesebroeck B, Dixit V, Thakar MS, Malarkannan S. Signaling by Fyn-ADAP via the Carma1-Bcl-10-MAP3K7 signalosome exclusively regulates inflammatory cytokine production in NK cells. Nat Immunol. 2013. 11/2013;14(11):1127–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fauriat C, Long EO, Ljunggren H-G, Bryceson YT. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood. 2010;115(11):2167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Freeman BE, Raué H-P, Hill AB, Slifka MK. Cytokine-mediated activation of NK cells during viral infection. J Virol. 2015;89(15):7922–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bregnard C, Guerra J, Dejardin S, Passalacqua F, Benkirane M, Laguette N. Upregulated LINE-1 activity in the Fanconi anemia cancer susceptibility syndrome leads to spontaneous pro-inflammatory cytokine production. EBioMedicine. 2016. Jun;8:184–94. PubMed PMID: 27428429. PMCID: PMC4919473. Epub 2016/07/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sivori S, Falco M, Della Chiesa M, Carlomagno S, Vitale M, Moretta L, Moretta A. CpG and double-stranded RNA trigger human NK cells by Toll-like receptors: Induction of cytokine release and cytotoxicity against tumors and dendritic cells. Proc Natl Acad Sci U S A. 2004. Jul 6;101(27):10116–21. PubMed PMID: 15218108. PMCID: PMC454174. Epub 2004/06/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chalifour A, Jeannin P, Gauchat JF, Blaecke A, Malissard M, N’Guyen T, Thieblemont N, Delneste Y. Direct bacterial protein PAMP recognition by human NK cells involves TLRs and triggers alpha-defensin production. Blood. 2004. Sep 15;104(6):1778–83. PubMed PMID: 15166032. Epub 2004/05/29. [DOI] [PubMed] [Google Scholar]
- 82.Lauzon NM, Mian F, MacKenzie R, Ashkar AA. The direct effects of Toll-like receptor ligands on human NK cell cytokine production and cytotoxicity. Cell Immunol. 2006. Jun;241(2):102–12. PubMed PMID: 17049504. Epub 2006/10/20. [DOI] [PubMed] [Google Scholar]
- 83.Sivori S, Carlomagno S, Moretta L, Moretta A. Comparison of different CpG oligodeoxynucleotide classes for their capability to stimulate human NK cells. Eur J Immunol. 2006. Apr;36(4):961–7. PubMed PMID: 16525994. Epub 2006/03/10. [DOI] [PubMed] [Google Scholar]
- 84.Sawaki J, Tsutsui H, Hayashi N, Yasuda K, Akira S, Tanizawa T, Nakanishi K. Type 1 cytokine/chemokine production by mouse NK cells following activation of their TLR/MyD88-mediated pathways. Int Immunol. 2007;19(3):311–20. [DOI] [PubMed] [Google Scholar]
- 85.Girart MV, Fuertes MB, Domaica CI, Rossi LE, Zwirner NW. Engagement of TLR3, TLR7, and NKG2D regulate IFN-gamma secretion but not NKG2D-mediated cytotoxicity by human NK cells stimulated with suboptimal doses of IL-12. J Immunol. 2007. Sep 15;179(6):3472–9. PubMed PMID: 17804388. Epub 2007/09/07. [DOI] [PubMed] [Google Scholar]