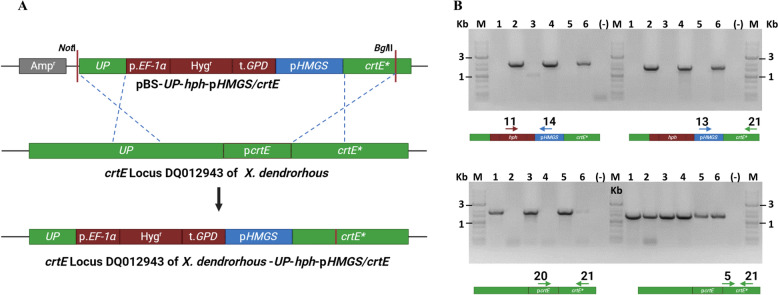
Fig. 2.
Representation of the integration of the Up-hph-pHMGS/crtE module in the X. dendrorhous genome. A Scheme illustrating the resulting product with the Up-hph-pHMGS/crtE module (digested with NotI and BglII enzymes), used to replace the native crtE gene promoter with the HMGS promoter through double homologous recombination. B PCR analysis of strains used in this work. A representation of the amplified fragment is provided beneath each gel, with primers indicated by numbered arrows according to Supplementary Table 1.2. Template DNA sources are as follows: CBS6938 (lane 1), CBS.pHMGS/crtE (lane 2), CBS.cyp61− (lane 3), CBS.cyp61−.pHMGS/crtE (lane 4), CBS.SRE1N.FLAG (lane 5), and CBS.SRE1N.FLAG.pHMGS/crtE (lane 6). The GeneRuler 1 kb Plus DNA Ladder (M) was used as a molecular weight marker, and (−) represents a negative control without DNA in the PCR reaction