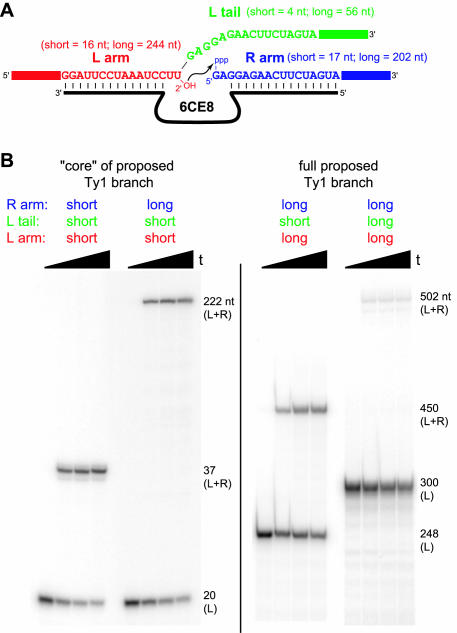
Figure 8.
Synthesis of the proposed Ty1 branched RNA by the 6CE8 deoxyribozyme. (A) Diagram and partial sequences of the proposed Ty1 branched RNA. In the specific set of long RNAs examined in this study, the L substrate is 300 nt (243 nt to the 5′-side of the branch site and 56 nt to the 3′-side of the branch site), and the R substrate is 202 nt. The minimal ‘core’ of this branched RNA consists solely of those nucleotides that bind directly to the 6CE8 DNA binding arms (shown explicitly here), plus 4 nt of the L tail. See Supplementary Material for complete RNA sequences and their correspondence to the various regions of the natural Ty1 RNA (U3, R, U5 and PBS). (B) Ligation by 6CE8 to form the minimal Ty1 core using various combinations of short and long L arm, L tail and R arm. For the L arm and R arm, ‘short’ refers to just the sequences shown explicitly in (A), and ‘long’ refers to the full sequences. For the L tail, ‘short’ refers to the 4 nt GAGG tail, and ‘long’ refers to 56 nt of sequence identical to the R arm. For all experiments, the L substrate was 5′-32P-radiolabeled; timepoints were taken at t = 0, 10, 20 and 30 min. The first two sets of lanes are 20% PAGE and the last two sets of lanes are 6% PAGE. Values of kobs and yield at 30 min timepoint: first set of lanes, 3.4 h−1 and 77%; second set of lanes, 3.2 h−1 and 61%; third set of lanes, 2.2 h−1 and 29% (50% at 90 min, data not shown); fourth set of lanes, ∼0.05 h−1 and 3%. For the first three sets of lanes, analogous experiments except with the short L tail sequence of AUCG instead of GAGG gave essentially equivalent results (data not shown).