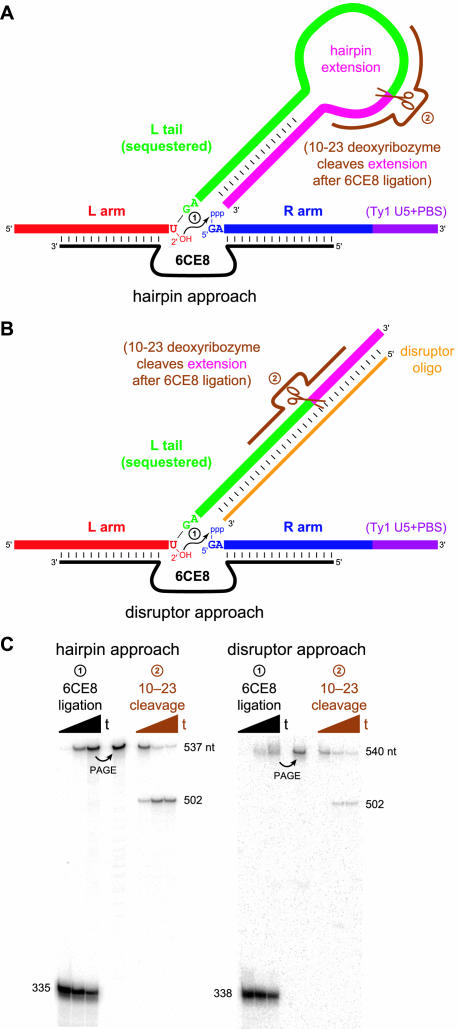
Figure 9.
The hairpin and disruptor approaches for synthesizing the full proposed Ty1 branched RNA. (A) Schematic diagram of the hairpin approach. The L tail (green) is extended with appropriate RNA sequence (pink) such that the tail is sequestered in a hairpin, thereby allowing the R arm (blue, with the same sequence as the L tail) to bind to the DNA (black). Following 6CE8-catalyzed branch formation, the extension sequence is cleaved by a 10–23 deoxyribozyme (brown). (B) Schematic diagram of the disruptor approach. The L tail is extended with arbitrary sequence (pink) that is different from the 3′-end of the R arm (purple). A disruptor DNA oligonucleotide (orange) then sequesters the L tail, and following 6CE8 ligation, the extension sequence is cleaved by a 10–23 deoxyribozyme (brown). See Supplementary Material for complete sequence information for both approaches, including several variants of the disruptor oligonucleotide; the optimal disruptor is not entirely complementary to the L tail region. (C) Experimental data that demonstrate successful application of both the hairpin and disruptor approaches. Sizes in nucleotides are shown along the side of each gel; see Supplementary Material for detailed sequence information. Timepoints were taken at t = 0, 5 and 30 min for 6CE8 ligation and 0, 10 and 30 min for 10–23 cleavage. For ligation using the hairpin approach, kobs = 2.6 h−1 with 44% yield at 30 min. For ligation using the disruptor approach, kobs = 2.3 h−1 with 25% yield at 30 min. The 10–23 cleavage yields at 30 min were 86% (hairpin approach) and 57% (disruptor approach).