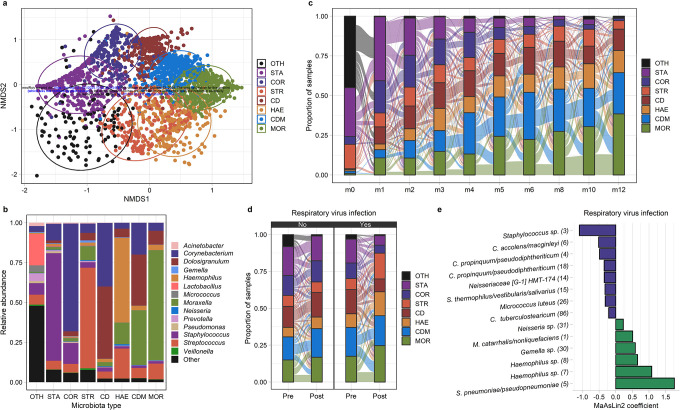
Figure 3. The dynamics of the upper respiratory microbiota among infants in Botswana.
a. NMDS plot based on Bray-Curtis distances depicts microbiota composition based on 16S rRNA sequencing of nasopharyngeal swabs collected from infants in Botswana during the first year of life. k-medoids clustering was used to identify 8 distinct microbiota types. Ellipses define the regions containing 80% of all samples that can be drawn from the underlying multivariate t distribution. b. Relative abundances of highly abundant genera in upper respiratory samples from infants (n=2,235 samples) by microbiota type. c.,d. Alluvial diagram depicting upper respiratory microbiota community state transitions during the first year of life (c.) and with respiratory virus infection (d.). e. MaAsLin2 was used to a fit generalized linear mixed model evaluating the association between respiratory virus infection and the relative abundances of ASVs within the infant upper respiratory microbiota. The coefficients from these models, which correspond to the relative effect sizes of associations, are shown for significant associations (q<0.20). ASVs that decreased/increased in abundance in association with respiratory virus infection are shown as blue/green bars. The taxonomic classification of each ASV based on BLAST searches is shown followed by a number in parentheses corresponding to the mean relative abundance of this ASV in infant nasopharyngeal samples (ASV1 was the most abundant ASV across all samples). URT microbiota types are abbreviated CD, Corynebacterium/Dolosigranulum-dominant; CDM, Corynebacterium/Dolosigranulum/Moraxella-dominant; COR, Corynebacterium-dominant; HAE, Haemophilus-dominant; MOR, Moraxella-dominant; STA, Staphylococcus-dominant; STR, Streptococcus-dominant; and OTH, “other.” ASV, amplicon sequence variant; non-metric multidimensional scaling