Abstract
The reverse transcriptase V207I mutation within the hepatitis B virus (HBV) polymerase is associated with resistance to lamivudine in vitro. The prevalence of this mutation in treatment-naïve patients was 1% (1/96). A follow-up of the patient carrying this mutation prior to treatment revealed no loss of sensitivity of HBV to lamivudine in vivo.
Treatment of chronic hepatitis B with lamivudine (3TC) leads to a substantial proportion of resistant hepatitis B virus (HBV) (3). Resistance-associated mutations outside the active site of the HBV polymerase (YMDD motif) include amino acid exchanges at the reverse transcriptase codon L180M (rtL180M) or the rtV207I codon (4, 7, 10). The 50% inhibitory concentration for HBV carrying the rtV207I mutation is 20 nM of 3TC, leading to a 3.6-fold resistance in vitro (4, 10). However, the relevance of this amino acid exchange to the resistance of HBV to 3TC in vivo is completely unknown.
We investigated 96 consecutive, chronic HBV carriers. HBV polymerase sequences were analyzed at baseline (n = 96), after 1 year (n = 96), and after 2 years of 3TC treatment (n = 92). Fifty patients carried HBV genotype A, and 46 were infected with HBV genotype D. The case patient was an immunocompetent male who was 19 years old at initiation of 3TC treatment. He carried HBV genotype A and was coinfected with hepatitis C virus (HCV) genotype 1a but tested negative for human immunodeficiency virus. The patient had received pegylated alpha interferon (pegIFN-α; 80 μg per week) and ribavirin (600 mg per day) 12 months prior to 3TC treatment due to his HCV infection without any effect on HBV DNA levels, concentration of serum hepatitis B surface antigen, or the status of hepatitis Be (HBe) antigen. He had never received 3TC or famciclovir before. At baseline of 3TC treatment (100 mg daily), his viral load was log10 8.3 HBV DNA copies/ml, while the HCV RNA level was below the detection limit of the PCR assay (lower log10 2 HCV RNA copies/ml). The HBe antigen test was positive, and the alanine aminotransferase (ALT) value at baseline was 20 U/ml (the normal value is below 22 U/ml).
Serum hepatitis surface antigen and HBe antigen levels were determined by enzyme-linked immunosorbent assay (Abbott, Wiesbaden, Germany). HBV copy numbers were quantified by real-time PCR (LightCycler-DNA Master SYBR Green; Roche Diagnostics, Basel, Switzerland) as described previously (8). Genotypic HBV resistance was analyzed by sequencing the HBV polymerase from codons rt103 to rt244 (1). Viral population kinetics were investigated by cloning amplicons of the HBV polymerase into Escherichia coli (TOPO-TA cloning system; Invitrogen, Karlsruhe, Germany). Fifteen clones were sequenced from each sample as described previously (11).
The prevalence of the rtV207I mutation in treatment-naïve patients was 1% (1/96). This patient was the case reported in this article. One additional patient carried wild-type HBV at baseline, but the rtV207I mutation emerged at month 24 of treatment in combination with the rtL180M and rtM204V exchanges. Therefore, the incidence of the rtV207I exchange was 0% (0/95) after 1 year and 1% (1/91) after 2 years of 3TC treatment (Table 1). In comparison, cumulative incidence rates were significantly higher for the rtL180M, rtM204I, and rtM204V mutations (Table 1).
TABLE 1.
Prevalence at baseline and incidence of resistance-associated mutations within the HBV polymerase during 3TC treatment as determined by direct sequencing of PCR products
| Mutation within HBV polymerase (rt) | Cumulative incidence rate during 3TC treatment (%) [no. with HBV genotype A/no. with HBV genotype D]
|
||
|---|---|---|---|
| At baseline (n = 96) | After 1 yr (n = 96) | After 2 yr (n = 92) | |
| L180M | 0 (0) | 10 (10) [8/2] | 26 (28) [19/7] |
| M204I (YIDD) | 0 (0) | 5 (5) [2/3] | 15 (16) [5/10] |
| M204V (YVDD) | 0 (0) | 14 (14) [14/0] | 26 (28) [21/5] |
| V207I | 1 (1)a | 0 (0)b | 1 (1) [1/0] |
This patient (case patient) carried HBV genotype A.
The patient with an rtV207I mutation at baseline (case patient) was excluded.
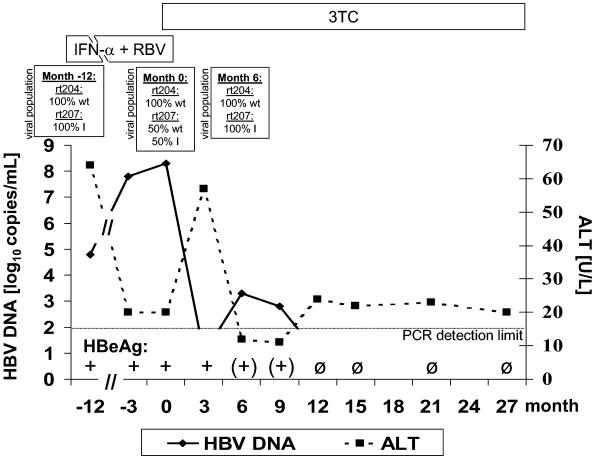
In the case patient, HBV population analysis showed a 100% rtV207I mutant virus at commencement of pegIFN-α-ribavirin treatment (Fig. 1). During this period of HCV-directed therapy, the concentration of HBV DNA increased from log10 4.8 to log10 8.3 HBV copies/ml after 12 months. At this time point, the HBV population consisted of 50% wild-type and 50% rtV207I mutant HBV. 3TC was added to pegIFN-α-ribavirin, resulting in a rapid decrease of HBV DNA to an undetectable HBV load. pegIFN-α-ribavirin was discontinued 1 month after initiation of 3TC therapy. The ALT concentration showed a self-limiting, short flair at month 3 of 3TC treatment. HBV DNA became detectable at months 6 and 9 (Fig. 1). Population analysis revealed a 100% rtV207I mutant HBV at month 6. From month 12 to month 27, HBV DNA was always below the detection limit of the PCR. HBe antigen was undetectable from month 12 on (Fig. 1), and anti-HBe antibodies developed.
FIG. 1.
Course of HBV DNA and ALT values for a patient with an rtV207I mutation at baseline of 3TC treatment. The proportions of wild-type and mutant HBV are depicted for specific time points. RBV, ribavirin; wt, wild-type HBV.
Apart from the rtV207I exchange, we found exclusively wild-type polymerase sequences in all clones derived from samples drawn at months −12, 0, and 6 of 3TC treatment, particularly within the YMDD motif (Fig. 1).
The patient had undetectable HCV RNA beginning at month 6 after initiation of pegIFN-α-ribavirin treatment and to 26 months after discontinuation. His ALT values were always in the normal range, apart from a short flair at month 3 of 3TC treatment.
The results of this study show that the rtV207I mutation is a very rare amino acid exchange and may represent a natural polymorphism. Without the selection pressure of antiviral treatment, the presence of a single rtV207I exchange led to suppression of wild-type HBV and predominance of the mutant strain. Therefore, the rtV207I mutant appears to be superior to wild-type HBV, possibly due to increased replication competence in vivo. As the mutation is very rare in chronic HBV carriers, however, this phenomenon cannot be generalized to all patients. We speculate that the mutant virus may have had an advantage over wild-type HBV because of inhibitory effects induced by HCV on HBV wild-type replication (2, 9).
After initiation of 3TC treatment, the rtV207I mutant was sensitive to this drug. This implies that the rtV207I exchange did not confer primary resistance of HBV to 3TC and that a single rtV207I mutation should not lead to exclusion of 3TC as an option for treatment of chronic hepatitis B. Our data also suggest that the rtV207I exchange has compensatory functions in vivo due to the finding that the rtV207I mutation was coselected in one patient in combination with the rtL180M/rtM204V resistance pattern. Characterization of the rtV207I mutation by transfection experiments showed a restoration of viral replication fitness in 3TC-resistant HBV exhibiting YMDD mutations (4). Hence, both the in vitro and our in vivo results corroborate the compensatory function of the rtV207I mutation. We cannot assess the in vivo relevance of the rtV207I mutation to the efficacy of famciclovir, because the case patient did not receive this drug during treatment.
It has been described that the overall prevalence of resistance-associated mutations in treatment-naïve, chronic HBV carriers ranges from 3.8% in Spain (6) to 28% in an Asian cohort (5). The results of our study clearly demonstrate that the impact of these naturally occurring HBV mutants on the outcome of antiviral treatment needs to be investigated in further clinical studies.
REFERENCES
- 1.Allen, M. I., M. Deslauriers, C. W. Andrews, G. A. Tipples, K.-A. Walters, D. L. Tyrell, N. Brown, Lamivudine Clinical Investigation Group, and L. D. Condreay. 1998. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Hepatology 27:1670-1677. [DOI] [PubMed] [Google Scholar]
- 2.Chen, S. Y., C. F. Kao, C. M. Chen, C. M. Shih, M. J. Hsu, C. H. Chao, S. H. Wang, L. R. You, and Y. H. W. Lee. 2003. Mechanisms for inhibition of hepatitis B virus gene expression and replication by hepatitis C virus core protein. J. Biol. Chem. 278:591-607. [DOI] [PubMed] [Google Scholar]
- 3.Dienstag, J. L., E. R. Schiff, T. L. Wright, P. R. Perillo, H. W. L. Hann, Z. Goodman, L. Crowther, L. D. Condreay, M. Woessner, M. Rubin, and N. A. Brown, et al. 1999. Lamivudine as initial treatment for chronic hepatitis B in the United States. N. Engl. J. Med. 341:1256-1263. [DOI] [PubMed] [Google Scholar]
- 4.Fu, L., and Y.-C. Cheng. 1998. Role of additional mutations outside the YMDD motif of hepatitis B virus polymerase in L(−)SddC (3TC) resistance. Biochem. Pharmacol. 55:1567-1572. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi, S., T. Ide, and M. Sata. 2001. Detection of YMDD motif mutations in some lamivudine-untreated asymptomatic hepatitis B virus carriers. J. Hepatol. 34:584-586. [DOI] [PubMed] [Google Scholar]
- 6.Leon, P., F. Pozo, and J. M. Echevarria. 2004. Detection of hepatitis B virus variants resistant to lamivudine and famciclovir among randomly selected chronic carriers from Spain. Enferm. Infecc. Microbiol. Clin. 22:133-137. [DOI] [PubMed] [Google Scholar]
- 7.Locarnini, S. 2003. Hepatitis B viral resistance: mechanisms and diagnosis. J. Hepatol. 39:S124-S132. [DOI] [PubMed] [Google Scholar]
- 8.Loeb, K. R., K. R. Jerome, J. Goddard, M. L. Huang, A. Cent, and L. Corey. 2000. High-throughput quantitative analysis of hepatitis B virus DNA in serum using the TaqMan fluorogenic detection method. Hepatology 32:626-629. [DOI] [PubMed] [Google Scholar]
- 9.Sagnelli, E., N. Coppola, C. Scolastico, P. Filippini, T. Santantonio, T. Stroffolini, and F. Piccinino. 2000. Virologic and clinical expressions of reciprocal inhibitory effect of hepatitis B, C, and delta viruses in patients with chronic hepatitis. Hepatology 32:1106-1110. [DOI] [PubMed] [Google Scholar]
- 10.Xiong, X., H. Yang, C. E. Westland, R. Zou, and C. S. Gibbs. 2000. In vitro evaluation of hepatitis B virus polymerase mutations associated with famciclovir resistance. Hepatology 31:219-224. [DOI] [PubMed] [Google Scholar]
- 11.Zöllner, B., J. Petersen, E. Puchhammer-Stöckl, J. Kletzmayr, M. Sterneck, L. Fischer, M. Schröter, R. Laufs, and H. H. Feucht. 2004. Viral features of lamivudine resistant hepatitis B virus genotypes A and D. Hepatology 39:42-50. [DOI] [PubMed] [Google Scholar]