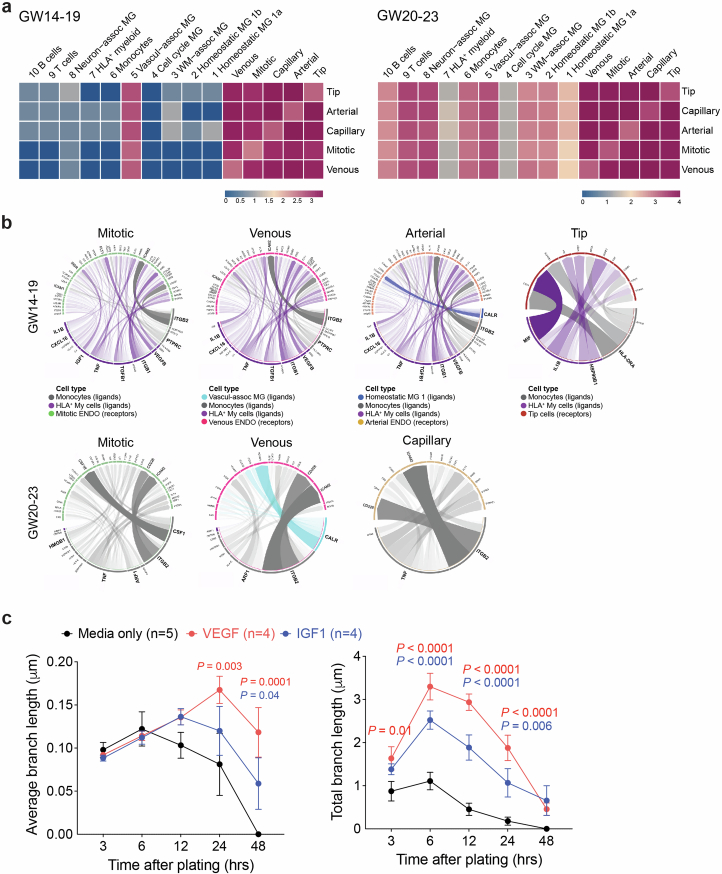
Extended Data Fig. 6. Bioinformatic analyses reveal potential signaling mechanisms that regulate the interactions between CD45+ immune cells and different subtypes of endothelial cells.
(a) CellphoneDB analyses reveal stage-dependent communications via ligand-receptor pairs between CD45+ subtypes and CD31+ endothelial subtypes at GW14-19 and GW20-23. (b) NicheNet analyses predict signaling pathways used by homeostatic microglia (c1a), HLA+ myeloid cells, monocytes, and VAM to interact with endothelial subtypes at GW14-19 and GW20-23. Data from panels a and b are from 2 independent biological samples at GW14-19, and 3 independent biological samples at GW20-23. ENDO, endothelial cells. MG, microglia. (c) Quantification of average and total endothelial branch lengths formed by HUVEC in Matrigel-based assays. The conditions include EGM-2 media alone or addition of VEGF or IGF1, which is one of the ligands used by CD45+ cells identified by NicheNet. Statistics use two-tailed, unpaired Student’s t-test, data represent mean ± SEM. The P values represent comparisons between HUVECs treated with VEGF or IGF1 vs no treatment. Not significant comparisons are not shown. n indicates the number of independent biological samples used for quantification.