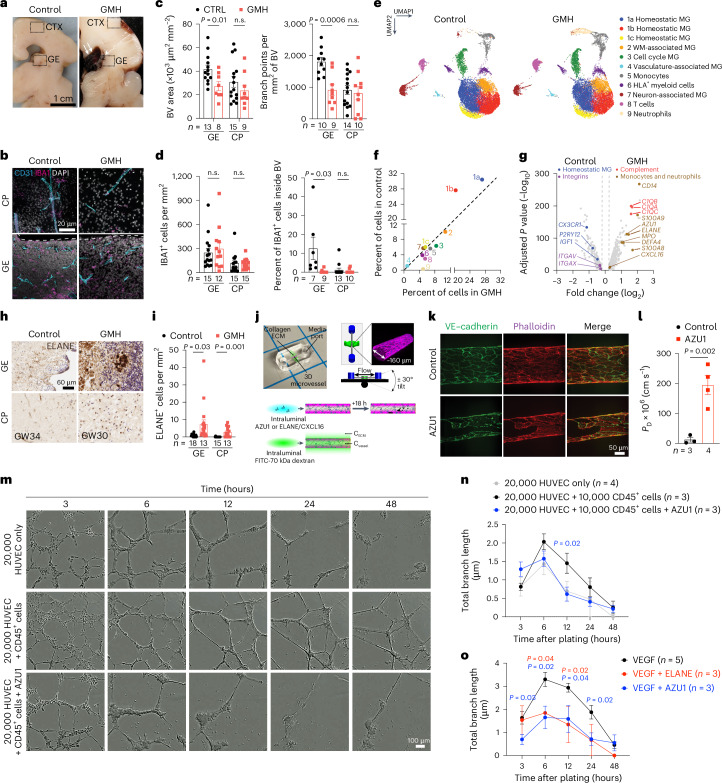
Fig. 6. Single-cell transcriptomics in CD45+ cells from GMH cases reveal activated neutrophils.
a, Gross images of a control prenatal brain (GW23) and a brain with GMH (GW24). b, IBA1+ cells and their relationship with CD31+ endothelial cells in the GE and cortical plate (CP) of control and GMH human brains. c, Quantification of densities of blood vessels and vascular branch points in the GE and CP of control and GMH cases. d, Quantification of IBA1+ cells and percentage of intravascular IBA1+ cells in the GE and CP of control and GMH cases. e, UMAP comparing CD45+ subtypes from GMH cases and age-matched control. MG, microglia; WM, white matter. f, A distribution plot comparing the relative abundance of CD45+ subtype in control versus GMH cases. g, A volcano plot showing DEGs and GO terms identified by pseudobulked scRNA-seq data in CD45+ cells from GMH versus control cases. The adjusted P values and fold changes were calculated using DESeq2. By default in DESeq2, the P values attained by the Wald test are corrected for multiple testing using the Benjamini–Hochberg method. The dashed lines indicate cutoffs for Pvalue of 0.05 and fold change of 1.2. The data from e, f and g are from two independent biological samples in each condition (control, GMH). h–i, Immunohistochemical stain for ELANE show increased number of neutrophils in the GE and CP of GMH cases. j, Experimental setup of in vitro vascular permeeability assay using 3D microfluidic microvessels. k, Fluorescence micrographs of VE–cadherin and actin in control and AZU1-treated microvessels. l, Quantification of vascular permeability in control and AZU1-treated microvessels. m, Images of HUVEC in Matrigel-based branching morphogenesis. n,o, Quantification of average and total endothelial branch lengths formed by HUVEC in Matrigel-based assays, showing neutrophil proteins (AZU1, ELANE) can suppress CD45+ cell-mediated (n) or VEGF-mediated (o) vascular morphogenesis. Statistics in c, d, i, l, n and o use a two-tailed, unpaired Student’s t-test, and the data represent the mean ± standard error of the mean. n.s., not significant. In n, the P values represent comparisons between HUVEC coincubated with CD45+ cells versus HUVEC only. In o, the P values represent comparisons between VEGF-primed HUVEC treated with AZU1 or ELANE versus VEGF-primed HUVEC only.