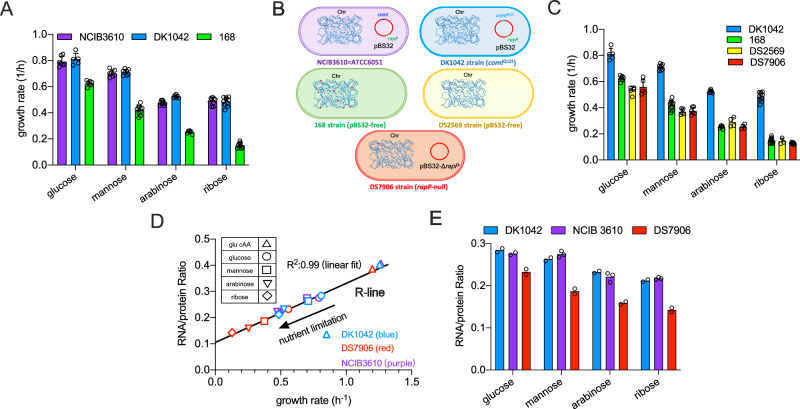
Fig. 1. The growth physiology of undomesticated and domesticated B. subtilis strains.
A The growth rates of NCIB 3610, DK1042, and 168 strains on various carbon sources. Data are presented as the mean values ± standard deviations (SD) of several biological replicates (for NCIB 3610 strain: n = 7 for glucose and mannose; n = 9 for arabinose and ribose; for DK1042 strain: n = 5, 8, 6, 10 for glucose, mannose, arabinose and ribose, respectively; for 168 strain: n = 7, 10, 6, 9 for glucose, mannose, arabinose and ribose, respectively). B Schematics of several B. subtilis strains used in this study. “Chr” is short for chromosome. NCIB 3610, DK1042, DS2569, and DS7906 share the same sequence of chromosomes while the domestical 168 strain carries additional mutations (represented by red points) in some chromosomal positions such as sfp, swrA, trpC, and gudB. C The growth rates of DK1042, 168, DS2569, and DS7906 strains on various carbon sources. Data of DK1042 and 168 are the same as (A). Data are presented as the mean values ± standard deviations (SD) of several biological replicates (for DS2569: n = 5, 5, 4, 3 for glucose, mannose, arabinose and ribose, respectively; for DS7906 strain: n = 5, 6, 4, 7 for glucose, mannose, arabinose and ribose, respectively). D Correlation of RNA/protein ratios versus growth rates for DK1042, NCIB 3610, and DS7906 strains under various nutrient conditions. Data of DK1042 and NCIB 3610 strains almost completely overlap with each other. E Comparison of RNA/protein ratios of DK1042, NCIB 3610, and DS7906 strains on different carbon sources. Individual data points are from different biological replicates. (n = 3 for NCIB 3610 on arabinose and ribose and n = 2 for all the rest conditions). Source data are provided as a Source Data file.