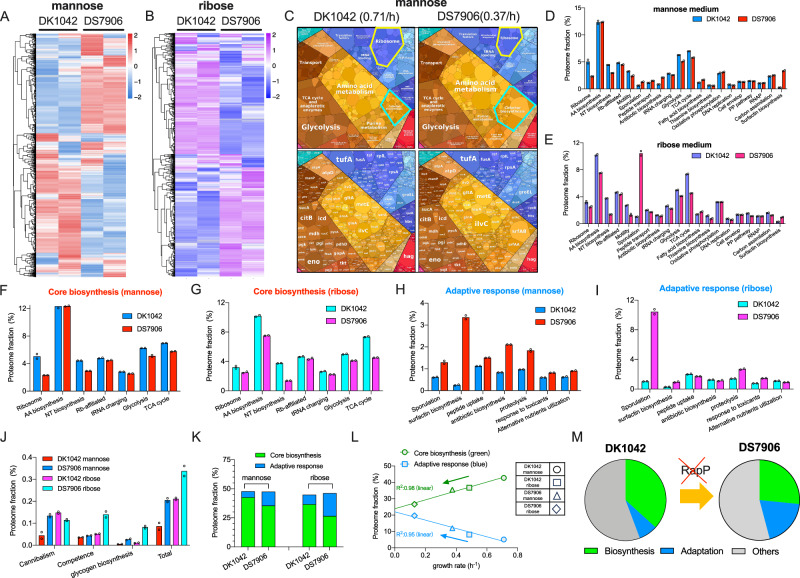
Fig. 4. The effect of RapP on proteome allocation of B. subtilis.
A, B Heatmaps of the proteomes of DK1042 and DS7906 strains growing in mannose and ribose media, respectively. C Proteome allocation of DK1042 and DS7906 strains visualized by the proteomaps website. Note that the term “mitochondrial biogenesis” inside of the large category “translation” was based on KEGG categorization consisting of both prokaryotes and eukaryotes. The readers should thus treat these proteins here as translation factors. Moreover, genes involved in biosynthesis of surfactin and some other lipopeptide antibiotics should be more properly treated as biosynthesis sector of secondary metabolites instead of cofactor biosynthesis. D, E The mass fractions of various functional proteome sectors of B. subtilis growing in mannose and ribose media. F, G The mass fractions of several core biosynthetic sectors of B. subtilis growing in mannose and ribose media. H, I The mass fractions of various adaptive response pathways of B. subtilis growing in mannose and ribose media. J The mass fractions of several low-abundant adaptive response pathways of B. subtilis. K The proteome fractions of total core biosynthetic pathways and total adaptive response pathways in mannose and ribose media. Total core biosynthetic sector refers to the sum of various sectors in (F) and (G) while total adaptive response sector refers to the sum of various sectors in (H–J). L The correlation of the proteome fractions of core biosynthetic category and adaptive response category with growth rates for the four conditions of B. subtilis. M Schematic illustration showing that the absence of RapP triggers the resource allocation from biosynthetic pathways to adaptive response pathways. For (D–J), individual data points correspond to two biological replicates (n = 2). Source data are provided as a Source Data file.