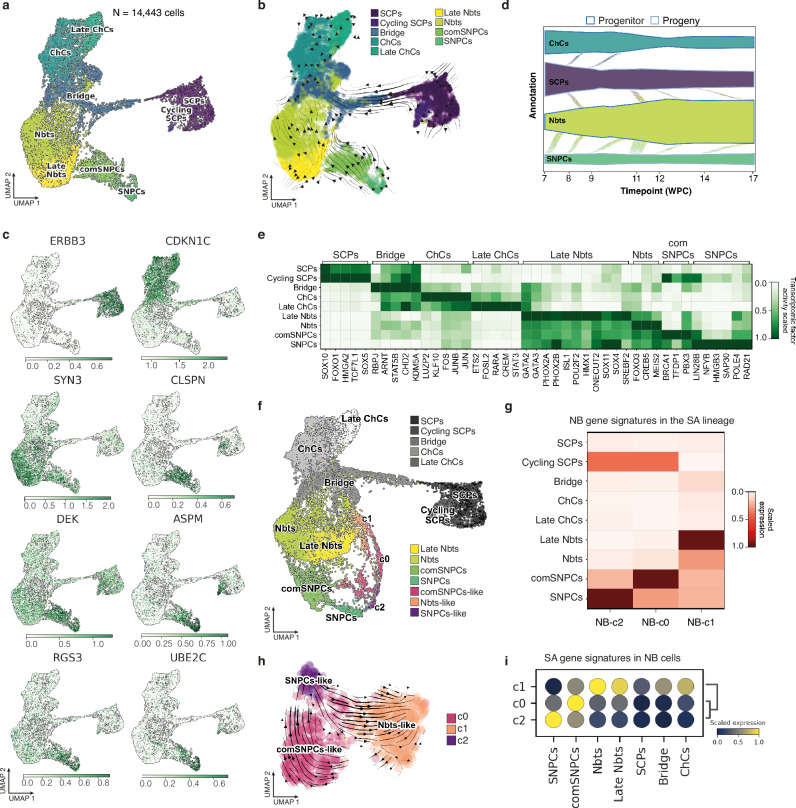
Fig. 2. NB cell plasticity in avian embryonic tissues relies on the adoption of features of the human SNPCs-to-neuroblasts differentiation program.
a UMAP embedding of 14,443 single cells of human physiological sympatho-adrenal cells colored by phenotypic state. The UMAP results from the integration of Jansky et al., 2021 and Kameneva et al., 2021 datasets. b UMAP embedding of human SA atlas with arrows highlighting the directions of state-to-state transitions. c Expression of key marker genes in UMAP embedding of the human SA atlas; color scales indicate log-normalized gene expression. d Probability mass flow as a function of time post-conception from either SCP or SNPC progenitor state using cellular growth and death score, combined with cell transcriptomic similarity to compute differentiation directions. e Heatmap of the scaled transcription factor activity for key genes in each physiological SA phenotypical state. Gene lists were extracted using differential expression with two-sided Wilcoxon rank sum test (p-value < 0.05, log fold change > 0.25, min.pct > 0.1). f Unimodal projection UMAP of NB cells -colored by transcriptomic states- mapped onto the human SA lineage atlas. g Heatmap of scaled expression values for NB-c0, NB-c1 and NB-c2 gene signature scores into cell populations of the SA lineage. NB-c0, NB-c1 and NB-c2 gene signatures were built from the intersection of NB gene markers grouped by phenotype (two-sided Wilcoxon rank sum test; p-value < 0.05, log fold change > 0.25, min.pct > 0.1) with anchor genes used for the unimodal projection UMAP shown in (f). h RNA velocity estimates for NB cells harvested at E14 on a UMAP embedding using dynamical modeling. i Dot plot illustrating SCPs-related or SNPCs-related differentiation dynamic gene signature scores into NB cell transcriptomic states. Cell cycle-related genes (listed in gene ontology GO:0007049 biological process) were removed from all signatures. Source data are provided as a source data file. ChCs Chromaffin Cells, Nbts Neuroblasts, SCPs Schwann Cell Precursors, SNPCs, Sympathetic Neuron Progenitor Cells, comSNPCs committed SNPCs.