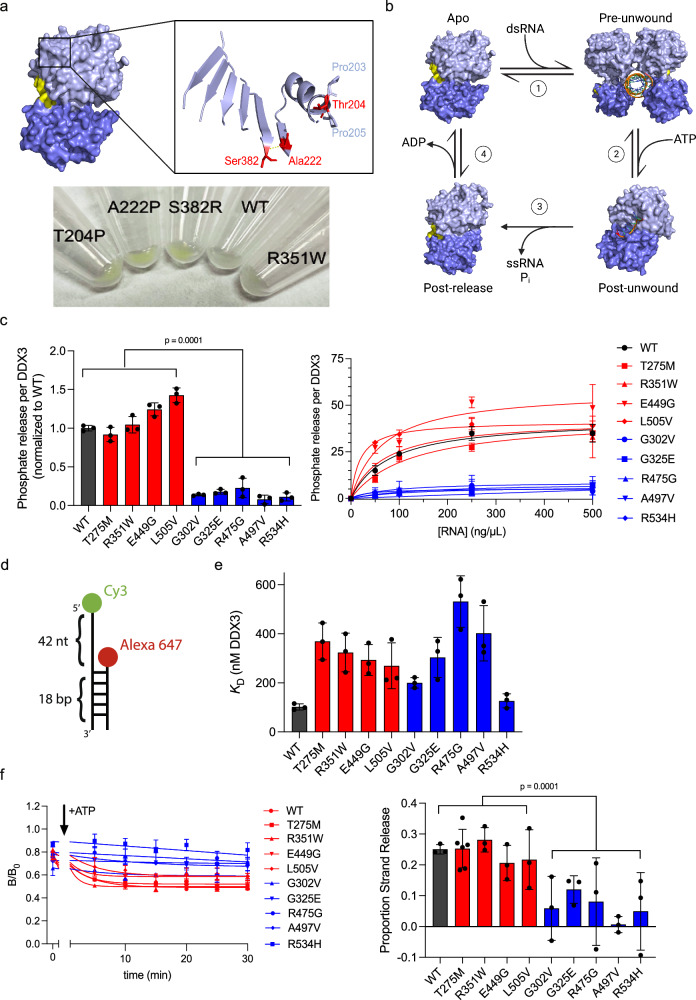
Fig. 2. Hollow mutants are characterized by their decreased ATPase and RNA strand release activities.
a Top, DDX3X apo structure zoomed into the three speckle mutants (red). Pro203 and Pro205 flanking Thr204 are indicated in light blue. The yellow line indicates the hydrogen bond between Ala222 and Ser382. Bottom, insoluble pellets of HeLa cells expressing mClover3-tagged WT DDX3X or the indicated mutants, after RIPA lysis and centrifugation. The green color of the speckle mutant pellets indicates their insolubility relative to WT DDX3X and R351W (a diffuse mutant). b Current model of the DDX3X helicase reaction. Light blue = N-terminal RecA-like domain, dark blue = the C-terminal RecA-like domain, orange = RNA. In the apo state (PDB 5E7I), a DDX3X dimer binds dsRNA (1) to form the pre-unwound state (PDB 6O5F). Upon addition of ATP, each monomer performs strand separation (2), converting to the post-unwound state (PDB 2DB3). Following ATP hydrolysis, DDX3X releases both the inorganic phosphate and the product ssRNA strand (3), forming the post-release state (PDB 4PXA). ADP release (4) returns DDX3X to apo state. c Left, ATPase activity of WT DDX3X or indicated mutants, measured as described in Methods, normalized to WT. Values represent means ± s.d., n = 3 replicates. Significance calculated using two-way ANOVA comparing diffuse mutants (and WT) to hollow mutants. Right, ATPase activities at the indicated concentration of total RNA. Values fit to Michaelis–Menten. Values = means ± s.d., n = 3 replicates. d Schematic of the dsRNA probe. e Summarized KD values for the indicated WT and mutant DDX3X. Values represent mean ± s.d., n = 3 independent reactions for WT and diffuse mutants, n = 6 for hollow mutants (n = 5 for G325E). f Left, time courses tracing the B/B0 (proportion strand release) for short strand RNA over, fit as described in Methods. Right, change in B/B0 signal from 0 to 30 min for each protein. Significance calculated using two-way ANOVA comparing diffuse mutants (and WT) to hollow mutants. Values represent means ± s.d., n = 3 independent replicates (n = 6 for T275M). Source data are provided as a Source Data file.